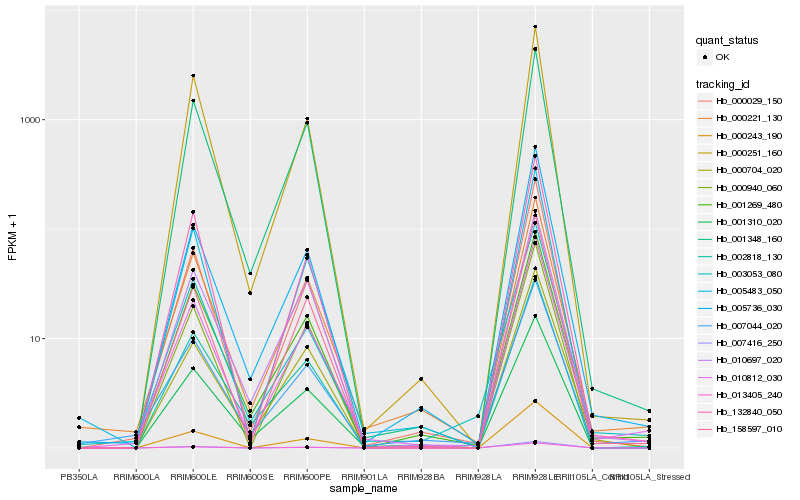
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_010812_030 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_000029_150 |
0.0615717225 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_158597_010 |
0.064277312 |
- |
- |
PREDICTED: serine hydroxymethyltransferase, mitochondrial [Jatropha curcas] |
| 4 |
Hb_000221_130 |
0.0647130079 |
- |
- |
thylakoid lumenal 29 kDa protein, chloroplastic [Jatropha curcas] |
| 5 |
Hb_001269_480 |
0.0673146561 |
- |
- |
PREDICTED: photosynthetic NDH subunit of subcomplex B 1, chloroplastic [Jatropha curcas] |
| 6 |
Hb_005483_050 |
0.0695188773 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_005736_030 |
0.0704061275 |
- |
- |
thioredoxin m(mitochondrial)-type, putative [Ricinus communis] |
| 8 |
Hb_132840_050 |
0.0736613915 |
- |
- |
PREDICTED: protein CURVATURE THYLAKOID 1B, chloroplastic [Jatropha curcas] |
| 9 |
Hb_000243_190 |
0.0738434866 |
transcription factor |
TF Family: MIKC |
flowering-related B-class MADS-box protein [Vitis vinifera] |
| 10 |
Hb_000940_060 |
0.0742769657 |
- |
- |
hypothetical protein POPTR_0009s13250g [Populus trichocarpa] |
| 11 |
Hb_007044_020 |
0.0757719838 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 12 |
Hb_001310_020 |
0.0775864959 |
- |
- |
PREDICTED: cyclin-D5-1-like isoform X2 [Jatropha curcas] |
| 13 |
Hb_002818_130 |
0.0798392313 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 14 |
Hb_000251_160 |
0.0818825465 |
- |
- |
chlorophyll A/B binding protein, putative [Ricinus communis] |
| 15 |
Hb_000704_020 |
0.0821699834 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_010697_020 |
0.082524351 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_003053_080 |
0.0831130679 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_013405_240 |
0.0853542336 |
- |
- |
Hydroxyproline-rich glycoprotein family protein [Theobroma cacao] |
| 19 |
Hb_001348_160 |
0.0865735351 |
- |
- |
Photosystem II 22 kDa protein, chloroplast precursor, putative [Ricinus communis] |
| 20 |
Hb_007416_250 |
0.0875367221 |
- |
- |
- |