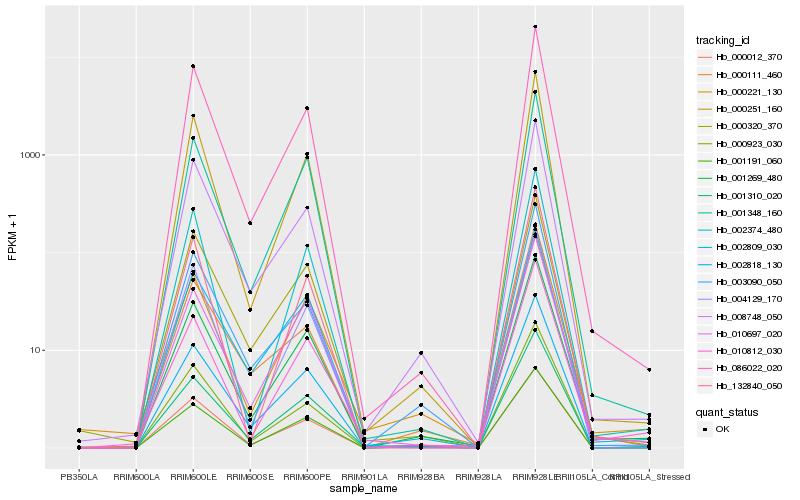
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001269_480 |
0.0 |
- |
- |
PREDICTED: photosynthetic NDH subunit of subcomplex B 1, chloroplastic [Jatropha curcas] |
| 2 |
Hb_001191_060 |
0.0499878369 |
- |
- |
PREDICTED: ankyrin repeat-containing protein At5g02620-like [Populus euphratica] |
| 3 |
Hb_002818_130 |
0.0513977039 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 4 |
Hb_001310_020 |
0.0548766212 |
- |
- |
PREDICTED: cyclin-D5-1-like isoform X2 [Jatropha curcas] |
| 5 |
Hb_000221_130 |
0.0570413184 |
- |
- |
thylakoid lumenal 29 kDa protein, chloroplastic [Jatropha curcas] |
| 6 |
Hb_001348_160 |
0.058302996 |
- |
- |
Photosystem II 22 kDa protein, chloroplast precursor, putative [Ricinus communis] |
| 7 |
Hb_086022_020 |
0.0589802873 |
- |
- |
RecName: Full=Ribulose bisphosphate carboxylase small chain, chloroplastic; Short=RuBisCO small subunit; Flags: Precursor [Hevea brasiliensis] |
| 8 |
Hb_000251_160 |
0.0601842187 |
- |
- |
chlorophyll A/B binding protein, putative [Ricinus communis] |
| 9 |
Hb_002809_030 |
0.0633160256 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_010697_020 |
0.0634778365 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_008748_050 |
0.0642808586 |
- |
- |
PREDICTED: chlorophyll a-b binding protein CP24 10A, chloroplastic-like [Populus euphratica] |
| 12 |
Hb_000923_030 |
0.065973554 |
- |
- |
F-box family protein [Populus trichocarpa] |
| 13 |
Hb_010812_030 |
0.0673146561 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_000012_370 |
0.0695291487 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_003090_050 |
0.0752256474 |
- |
- |
fructose-1,6-bisphosphatase, putative [Ricinus communis] |
| 16 |
Hb_002374_480 |
0.0752353757 |
- |
- |
Photosystem II 22 kDa protein, chloroplast precursor, putative [Ricinus communis] |
| 17 |
Hb_004129_170 |
0.0756006807 |
- |
- |
PREDICTED: protein trichome birefringence-like 43 [Jatropha curcas] |
| 18 |
Hb_000320_370 |
0.0756339098 |
- |
- |
PREDICTED: ATP synthase gamma chain, chloroplastic [Jatropha curcas] |
| 19 |
Hb_000111_460 |
0.0765940394 |
- |
- |
PREDICTED: uncharacterized protein LOC105631266 [Jatropha curcas] |
| 20 |
Hb_132840_050 |
0.0783386808 |
- |
- |
PREDICTED: protein CURVATURE THYLAKOID 1B, chloroplastic [Jatropha curcas] |