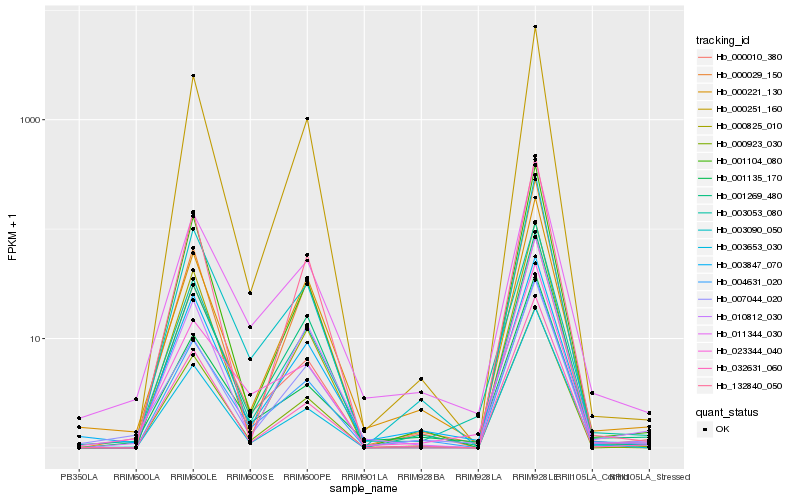
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003053_080 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_007044_020 |
0.0750366017 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 3 |
Hb_032631_060 |
0.0754712312 |
- |
- |
Myosin heavy chain, putative [Ricinus communis] |
| 4 |
Hb_000923_030 |
0.0755664069 |
- |
- |
F-box family protein [Populus trichocarpa] |
| 5 |
Hb_001135_170 |
0.0767300155 |
transcription factor |
TF Family: Orphans |
Two-component response regulator ARR2, putative [Ricinus communis] |
| 6 |
Hb_011344_030 |
0.0782061383 |
- |
- |
PREDICTED: elongation factor G-2, chloroplastic [Jatropha curcas] |
| 7 |
Hb_001104_080 |
0.0785376569 |
- |
- |
peroxiredoxin, putative [Ricinus communis] |
| 8 |
Hb_004631_020 |
0.0794658624 |
- |
- |
PREDICTED: protein TRANSPARENT TESTA 12-like [Jatropha curcas] |
| 9 |
Hb_001269_480 |
0.0814534267 |
- |
- |
PREDICTED: photosynthetic NDH subunit of subcomplex B 1, chloroplastic [Jatropha curcas] |
| 10 |
Hb_010812_030 |
0.0831130679 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_003653_030 |
0.0848607167 |
- |
- |
nucleic acid binding protein, putative [Ricinus communis] |
| 12 |
Hb_000825_010 |
0.0848790183 |
- |
- |
PREDICTED: trans-cinnamate 4-monooxygenase [Jatropha curcas] |
| 13 |
Hb_000029_150 |
0.0850334377 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_003090_050 |
0.0871777907 |
- |
- |
fructose-1,6-bisphosphatase, putative [Ricinus communis] |
| 15 |
Hb_132840_050 |
0.0883392484 |
- |
- |
PREDICTED: protein CURVATURE THYLAKOID 1B, chloroplastic [Jatropha curcas] |
| 16 |
Hb_000251_160 |
0.0900017127 |
- |
- |
chlorophyll A/B binding protein, putative [Ricinus communis] |
| 17 |
Hb_000221_130 |
0.0908078697 |
- |
- |
thylakoid lumenal 29 kDa protein, chloroplastic [Jatropha curcas] |
| 18 |
Hb_003847_070 |
0.0909094314 |
- |
- |
PREDICTED: violaxanthin de-epoxidase, chloroplastic [Jatropha curcas] |
| 19 |
Hb_000010_380 |
0.091869336 |
- |
- |
hypothetical protein CICLE_v10020897mg [Citrus clementina] |
| 20 |
Hb_023344_040 |
0.0919914051 |
- |
- |
conserved hypothetical protein [Ricinus communis] |