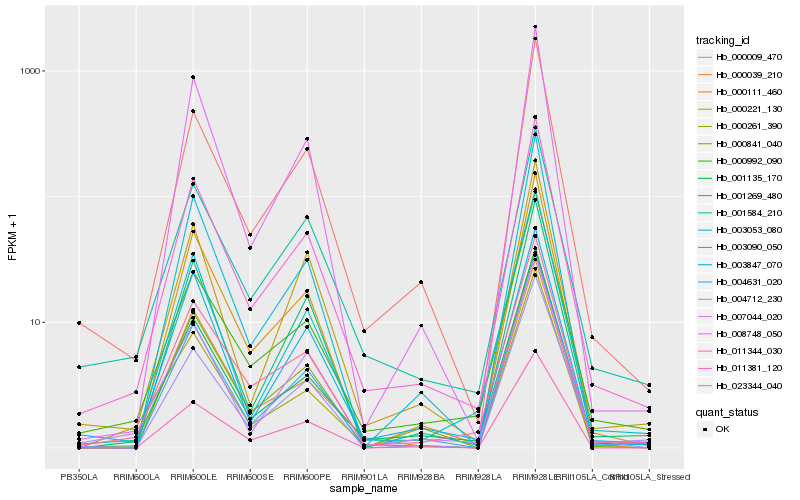
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_011344_030 |
0.0 |
- |
- |
PREDICTED: elongation factor G-2, chloroplastic [Jatropha curcas] |
| 2 |
Hb_000009_470 |
0.0563377649 |
- |
- |
Oxygen-evolving enhancer protein 1, chloroplast precursor, putative [Ricinus communis] |
| 3 |
Hb_023344_040 |
0.0568355221 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_000111_460 |
0.066834741 |
- |
- |
PREDICTED: uncharacterized protein LOC105631266 [Jatropha curcas] |
| 5 |
Hb_001135_170 |
0.0673517763 |
transcription factor |
TF Family: Orphans |
Two-component response regulator ARR2, putative [Ricinus communis] |
| 6 |
Hb_007044_020 |
0.0727123959 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 7 |
Hb_000992_090 |
0.0746659396 |
transcription factor |
TF Family: Orphans |
CIL, putative [Ricinus communis] |
| 8 |
Hb_003053_080 |
0.0782061383 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_004631_020 |
0.0789987276 |
- |
- |
PREDICTED: protein TRANSPARENT TESTA 12-like [Jatropha curcas] |
| 10 |
Hb_003847_070 |
0.07921744 |
- |
- |
PREDICTED: violaxanthin de-epoxidase, chloroplastic [Jatropha curcas] |
| 11 |
Hb_003090_050 |
0.0829807007 |
- |
- |
fructose-1,6-bisphosphatase, putative [Ricinus communis] |
| 12 |
Hb_008748_050 |
0.0848069793 |
- |
- |
PREDICTED: chlorophyll a-b binding protein CP24 10A, chloroplastic-like [Populus euphratica] |
| 13 |
Hb_001584_210 |
0.0851053807 |
- |
- |
PREDICTED: chloroplast stem-loop binding protein of 41 kDa b, chloroplastic [Jatropha curcas] |
| 14 |
Hb_000841_040 |
0.0873450826 |
transcription factor |
TF Family: C2C2-CO-like |
transcription factor, putative [Ricinus communis] |
| 15 |
Hb_000221_130 |
0.0876974502 |
- |
- |
thylakoid lumenal 29 kDa protein, chloroplastic [Jatropha curcas] |
| 16 |
Hb_001269_480 |
0.088152294 |
- |
- |
PREDICTED: photosynthetic NDH subunit of subcomplex B 1, chloroplastic [Jatropha curcas] |
| 17 |
Hb_000039_210 |
0.0883201264 |
- |
- |
PREDICTED: uncharacterized protein LOC105649449 [Jatropha curcas] |
| 18 |
Hb_011381_120 |
0.0903702534 |
- |
- |
PREDICTED: protein ASPARTIC PROTEASE IN GUARD CELL 2-like [Jatropha curcas] |
| 19 |
Hb_000261_390 |
0.0905707356 |
transcription factor |
TF Family: ARID |
PREDICTED: high mobility group B protein 15 [Jatropha curcas] |
| 20 |
Hb_004712_230 |
0.0913760847 |
- |
- |
PREDICTED: retrotransposon-like protein 1 [Jatropha curcas] |