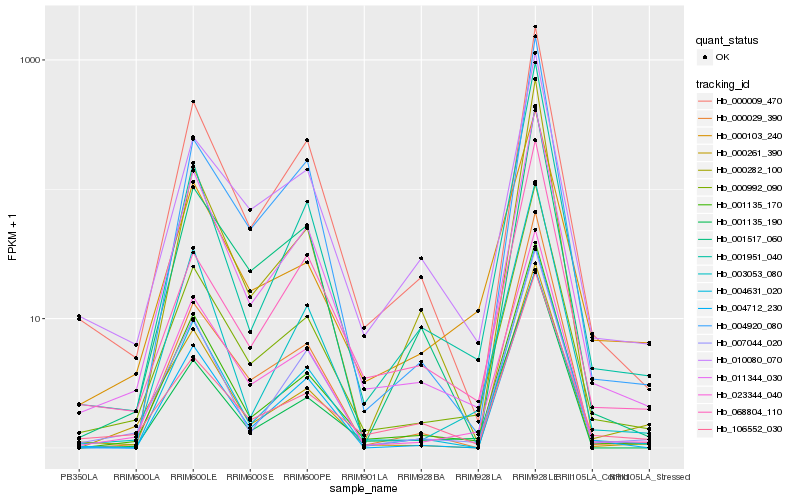
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000992_090 |
0.0 |
transcription factor |
TF Family: Orphans |
CIL, putative [Ricinus communis] |
| 2 |
Hb_023344_040 |
0.0620150176 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_001135_170 |
0.0656222935 |
transcription factor |
TF Family: Orphans |
Two-component response regulator ARR2, putative [Ricinus communis] |
| 4 |
Hb_011344_030 |
0.0746659396 |
- |
- |
PREDICTED: elongation factor G-2, chloroplastic [Jatropha curcas] |
| 5 |
Hb_000009_470 |
0.0815986298 |
- |
- |
Oxygen-evolving enhancer protein 1, chloroplast precursor, putative [Ricinus communis] |
| 6 |
Hb_000029_390 |
0.0859590125 |
transcription factor |
TF Family: mTERF |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_000103_240 |
0.08709407 |
- |
- |
PREDICTED: 1,2-dihydroxy-3-keto-5-methylthiopentene dioxygenase 1 [Jatropha curcas] |
| 8 |
Hb_010080_070 |
0.0896302575 |
- |
- |
PREDICTED: ATP-dependent zinc metalloprotease FTSH, chloroplastic [Jatropha curcas] |
| 9 |
Hb_001951_040 |
0.0900770103 |
- |
- |
PREDICTED: probable starch synthase 4, chloroplastic/amyloplastic isoform X1 [Jatropha curcas] |
| 10 |
Hb_004631_020 |
0.0903028592 |
- |
- |
PREDICTED: protein TRANSPARENT TESTA 12-like [Jatropha curcas] |
| 11 |
Hb_106552_030 |
0.0939181653 |
- |
- |
hypothetical protein POPTR_0006s23180g [Populus trichocarpa] |
| 12 |
Hb_007044_020 |
0.0944962782 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 13 |
Hb_001135_190 |
0.0967188745 |
transcription factor |
TF Family: Orphans |
Pseudo response regulator isoform 11 [Theobroma cacao] |
| 14 |
Hb_004712_230 |
0.0980954363 |
- |
- |
PREDICTED: retrotransposon-like protein 1 [Jatropha curcas] |
| 15 |
Hb_000261_390 |
0.0988324142 |
transcription factor |
TF Family: ARID |
PREDICTED: high mobility group B protein 15 [Jatropha curcas] |
| 16 |
Hb_001517_060 |
0.0992013631 |
- |
- |
phosphoribulose kinase, putative [Ricinus communis] |
| 17 |
Hb_003053_080 |
0.1011872478 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_004920_080 |
0.1032836322 |
- |
- |
PREDICTED: calvin cycle protein CP12-2, chloroplastic-like [Jatropha curcas] |
| 19 |
Hb_000282_100 |
0.1057102456 |
- |
- |
PREDICTED: glycine dehydrogenase (decarboxylating), mitochondrial [Jatropha curcas] |
| 20 |
Hb_068804_110 |
0.1067423161 |
- |
- |
PREDICTED: serine hydroxymethyltransferase, mitochondrial [Jatropha curcas] |