| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
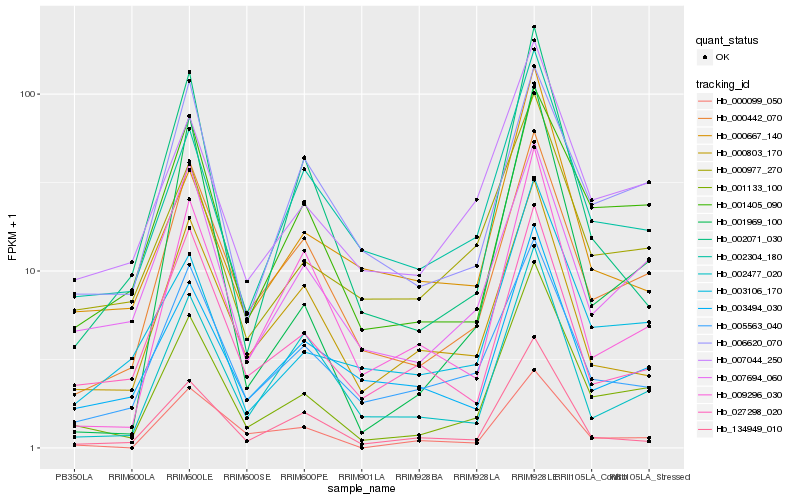
Hb_001133_100 |
0.0 |
- |
- |
PREDICTED: oligopeptide transporter 6-like [Jatropha curcas] |
| 2 |
Hb_000442_070 |
0.1366023916 |
- |
- |
PREDICTED: uncharacterized protein LOC105645538 [Jatropha curcas] |
| 3 |
Hb_007044_250 |
0.1372571889 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_001405_090 |
0.143451554 |
- |
- |
PREDICTED: photosynthetic NDH subunit of subcomplex B 5, chloroplastic [Jatropha curcas] |
| 5 |
Hb_003106_170 |
0.1465944935 |
- |
- |
Endonuclease III, putative [Ricinus communis] |
| 6 |
Hb_007694_060 |
0.1520799831 |
- |
- |
PREDICTED: 50S ribosomal protein L35, chloroplastic [Jatropha curcas] |
| 7 |
Hb_000977_270 |
0.1527538483 |
- |
- |
PREDICTED: phosphoglycolate phosphatase 1B, chloroplastic [Jatropha curcas] |
| 8 |
Hb_134949_010 |
0.1533513 |
- |
- |
PREDICTED: uncharacterized protein LOC105629574 [Jatropha curcas] |
| 9 |
Hb_002304_180 |
0.1579224001 |
- |
- |
PREDICTED: magnesium-chelatase subunit ChlI, chloroplastic [Jatropha curcas] |
| 10 |
Hb_001969_100 |
0.1622626141 |
- |
- |
signal transducer, putative [Ricinus communis] |
| 11 |
Hb_003494_030 |
0.1628326457 |
- |
- |
PREDICTED: protease Do-like 8, chloroplastic [Jatropha curcas] |
| 12 |
Hb_009296_030 |
0.1636819039 |
- |
- |
PREDICTED: uncharacterized protein LOC105634328 [Jatropha curcas] |
| 13 |
Hb_002477_020 |
0.1640824544 |
- |
- |
PREDICTED: uncharacterized protein LOC105631402 [Jatropha curcas] |
| 14 |
Hb_005563_040 |
0.1648065517 |
- |
- |
PREDICTED: uncharacterized protein LOC105630105 isoform X1 [Jatropha curcas] |
| 15 |
Hb_000099_050 |
0.1663826334 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_002071_030 |
0.1682468687 |
- |
- |
PREDICTED: ACT domain-containing protein ACR11-like isoform X1 [Gossypium raimondii] |
| 17 |
Hb_000803_170 |
0.1706956971 |
- |
- |
PREDICTED: uncharacterized protein LOC105648312 [Jatropha curcas] |
| 18 |
Hb_027298_020 |
0.1720169758 |
- |
- |
hypothetical protein POPTR_0005s05760g [Populus trichocarpa] |
| 19 |
Hb_006620_070 |
0.1756399369 |
- |
- |
chloroplast 30S ribosomal protein S13 [Hevea brasiliensis] |
| 20 |
Hb_000667_140 |
0.1774994468 |
- |
- |
PREDICTED: uridine kinase-like protein 3 isoform X1 [Jatropha curcas] |