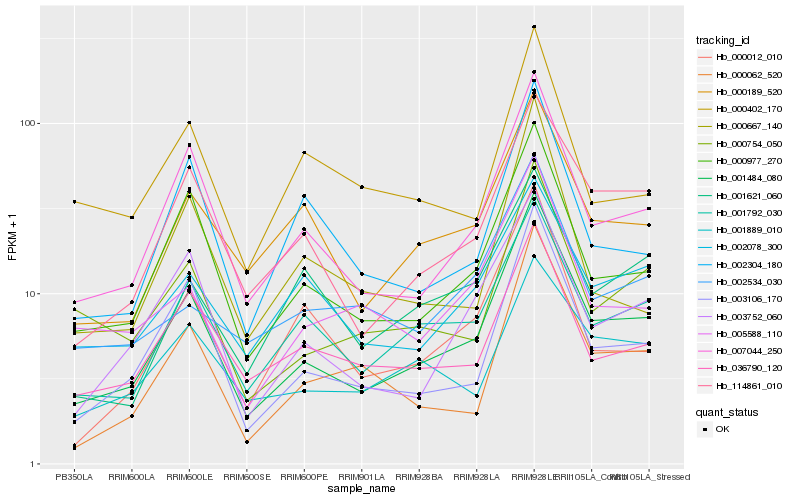
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001484_080 |
0.0 |
- |
- |
2-deoxyglucose-6-phosphate phosphatase, putative [Ricinus communis] |
| 2 |
Hb_007044_250 |
0.0919015937 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_000977_270 |
0.1022787666 |
- |
- |
PREDICTED: phosphoglycolate phosphatase 1B, chloroplastic [Jatropha curcas] |
| 4 |
Hb_003106_170 |
0.1050827789 |
- |
- |
Endonuclease III, putative [Ricinus communis] |
| 5 |
Hb_114861_010 |
0.107566401 |
- |
- |
STRESS ENHANCED protein 1 [Populus trichocarpa] |
| 6 |
Hb_001792_030 |
0.1157686939 |
- |
- |
PREDICTED: aminoacyl tRNA synthase complex-interacting multifunctional protein 1 [Jatropha curcas] |
| 7 |
Hb_003752_060 |
0.1279474219 |
- |
- |
PREDICTED: epoxide hydrolase 3 [Jatropha curcas] |
| 8 |
Hb_000189_520 |
0.128014488 |
- |
- |
PREDICTED: uncharacterized protein LOC105647658 [Jatropha curcas] |
| 9 |
Hb_000754_050 |
0.1310413453 |
- |
- |
PREDICTED: thylakoid lumenal 19 kDa protein, chloroplastic [Jatropha curcas] |
| 10 |
Hb_000062_520 |
0.1327470449 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_036790_120 |
0.1350083278 |
- |
- |
PREDICTED: uncharacterized protein LOC105645122 isoform X1 [Jatropha curcas] |
| 12 |
Hb_005588_110 |
0.1431121865 |
- |
- |
hypothetical protein POPTR_0002s12860g [Populus trichocarpa] |
| 13 |
Hb_000012_010 |
0.1440522302 |
- |
- |
hypothetical protein JCGZ_07709 [Jatropha curcas] |
| 14 |
Hb_002304_180 |
0.1518463075 |
- |
- |
PREDICTED: magnesium-chelatase subunit ChlI, chloroplastic [Jatropha curcas] |
| 15 |
Hb_001889_010 |
0.1520381281 |
transcription factor |
TF Family: Orphans |
PREDICTED: protein CHLOROPLAST IMPORT APPARATUS 2 [Jatropha curcas] |
| 16 |
Hb_002534_030 |
0.1524848237 |
transcription factor |
TF Family: M-type |
mads box protein, putative [Ricinus communis] |
| 17 |
Hb_002078_300 |
0.1556380722 |
- |
- |
PREDICTED: uncharacterized protein LOC105644817 isoform X1 [Jatropha curcas] |
| 18 |
Hb_001621_060 |
0.1604854075 |
- |
- |
PREDICTED: thylakoid lumenal 15 kDa protein 1, chloroplastic [Jatropha curcas] |
| 19 |
Hb_000667_140 |
0.1621563173 |
- |
- |
PREDICTED: uridine kinase-like protein 3 isoform X1 [Jatropha curcas] |
| 20 |
Hb_000402_170 |
0.164091331 |
- |
- |
PREDICTED: glutamate--glyoxylate aminotransferase 2 [Jatropha curcas] |