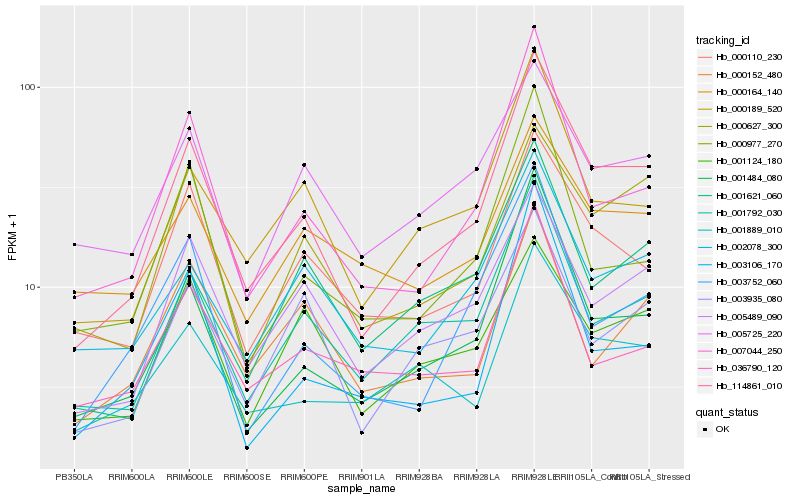
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_114861_010 |
0.0 |
- |
- |
STRESS ENHANCED protein 1 [Populus trichocarpa] |
| 2 |
Hb_001484_080 |
0.107566401 |
- |
- |
2-deoxyglucose-6-phosphate phosphatase, putative [Ricinus communis] |
| 3 |
Hb_001792_030 |
0.1160560705 |
- |
- |
PREDICTED: aminoacyl tRNA synthase complex-interacting multifunctional protein 1 [Jatropha curcas] |
| 4 |
Hb_007044_250 |
0.116394643 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_000189_520 |
0.1208351819 |
- |
- |
PREDICTED: uncharacterized protein LOC105647658 [Jatropha curcas] |
| 6 |
Hb_005489_090 |
0.1240093101 |
- |
- |
PREDICTED: peptidyl-tRNA hydrolase ICT1, mitochondrial [Jatropha curcas] |
| 7 |
Hb_000110_230 |
0.1282229621 |
- |
- |
Plastid-specific 30S ribosomal protein 3, chloroplast precursor, putative [Ricinus communis] |
| 8 |
Hb_002078_300 |
0.1360933479 |
- |
- |
PREDICTED: uncharacterized protein LOC105644817 isoform X1 [Jatropha curcas] |
| 9 |
Hb_003935_080 |
0.1361029197 |
- |
- |
PREDICTED: mechanosensitive ion channel protein 2, chloroplastic [Jatropha curcas] |
| 10 |
Hb_000627_300 |
0.1361739894 |
- |
- |
PREDICTED: uncharacterized protein LOC105632672 [Jatropha curcas] |
| 11 |
Hb_001889_010 |
0.1373429442 |
transcription factor |
TF Family: Orphans |
PREDICTED: protein CHLOROPLAST IMPORT APPARATUS 2 [Jatropha curcas] |
| 12 |
Hb_003752_060 |
0.1414536282 |
- |
- |
PREDICTED: epoxide hydrolase 3 [Jatropha curcas] |
| 13 |
Hb_000977_270 |
0.1436389499 |
- |
- |
PREDICTED: phosphoglycolate phosphatase 1B, chloroplastic [Jatropha curcas] |
| 14 |
Hb_005725_220 |
0.1466592763 |
- |
- |
PREDICTED: uncharacterized protein LOC105630808 [Jatropha curcas] |
| 15 |
Hb_001124_180 |
0.1504543616 |
- |
- |
PREDICTED: uncharacterized protein LOC105171432 [Sesamum indicum] |
| 16 |
Hb_001621_060 |
0.1509795695 |
- |
- |
PREDICTED: thylakoid lumenal 15 kDa protein 1, chloroplastic [Jatropha curcas] |
| 17 |
Hb_036790_120 |
0.1519929531 |
- |
- |
PREDICTED: uncharacterized protein LOC105645122 isoform X1 [Jatropha curcas] |
| 18 |
Hb_000152_480 |
0.1544371398 |
- |
- |
PREDICTED: protein LOW PSII ACCUMULATION 2, chloroplastic [Jatropha curcas] |
| 19 |
Hb_003106_170 |
0.1559514813 |
- |
- |
Endonuclease III, putative [Ricinus communis] |
| 20 |
Hb_000164_140 |
0.1576372652 |
- |
- |
cinnamoyl-CoA reductase, putative [Ricinus communis] |