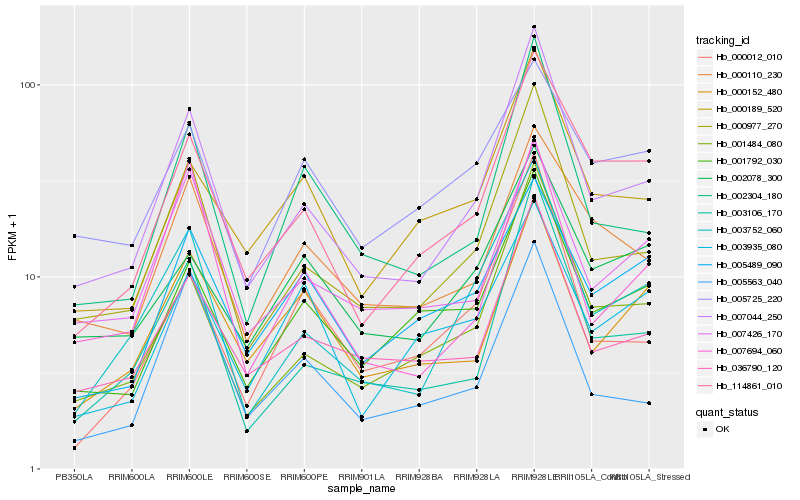
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003752_060 |
0.0 |
- |
- |
PREDICTED: epoxide hydrolase 3 [Jatropha curcas] |
| 2 |
Hb_007044_250 |
0.0952339669 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_000977_270 |
0.1055872188 |
- |
- |
PREDICTED: phosphoglycolate phosphatase 1B, chloroplastic [Jatropha curcas] |
| 4 |
Hb_001484_080 |
0.1279474219 |
- |
- |
2-deoxyglucose-6-phosphate phosphatase, putative [Ricinus communis] |
| 5 |
Hb_003106_170 |
0.1336666096 |
- |
- |
Endonuclease III, putative [Ricinus communis] |
| 6 |
Hb_114861_010 |
0.1414536282 |
- |
- |
STRESS ENHANCED protein 1 [Populus trichocarpa] |
| 7 |
Hb_036790_120 |
0.1640317159 |
- |
- |
PREDICTED: uncharacterized protein LOC105645122 isoform X1 [Jatropha curcas] |
| 8 |
Hb_001792_030 |
0.1658537568 |
- |
- |
PREDICTED: aminoacyl tRNA synthase complex-interacting multifunctional protein 1 [Jatropha curcas] |
| 9 |
Hb_007694_060 |
0.1671012174 |
- |
- |
PREDICTED: 50S ribosomal protein L35, chloroplastic [Jatropha curcas] |
| 10 |
Hb_000012_010 |
0.1700784534 |
- |
- |
hypothetical protein JCGZ_07709 [Jatropha curcas] |
| 11 |
Hb_007426_170 |
0.1712647858 |
transcription factor |
TF Family: TCP |
- |
| 12 |
Hb_002304_180 |
0.1721599151 |
- |
- |
PREDICTED: magnesium-chelatase subunit ChlI, chloroplastic [Jatropha curcas] |
| 13 |
Hb_002078_300 |
0.1743132945 |
- |
- |
PREDICTED: uncharacterized protein LOC105644817 isoform X1 [Jatropha curcas] |
| 14 |
Hb_005489_090 |
0.1796371102 |
- |
- |
PREDICTED: peptidyl-tRNA hydrolase ICT1, mitochondrial [Jatropha curcas] |
| 15 |
Hb_000110_230 |
0.179766227 |
- |
- |
Plastid-specific 30S ribosomal protein 3, chloroplast precursor, putative [Ricinus communis] |
| 16 |
Hb_005725_220 |
0.1831005261 |
- |
- |
PREDICTED: uncharacterized protein LOC105630808 [Jatropha curcas] |
| 17 |
Hb_000189_520 |
0.1840727324 |
- |
- |
PREDICTED: uncharacterized protein LOC105647658 [Jatropha curcas] |
| 18 |
Hb_000152_480 |
0.1859446956 |
- |
- |
PREDICTED: protein LOW PSII ACCUMULATION 2, chloroplastic [Jatropha curcas] |
| 19 |
Hb_005563_040 |
0.188919421 |
- |
- |
PREDICTED: uncharacterized protein LOC105630105 isoform X1 [Jatropha curcas] |
| 20 |
Hb_003935_080 |
0.1905302898 |
- |
- |
PREDICTED: mechanosensitive ion channel protein 2, chloroplastic [Jatropha curcas] |