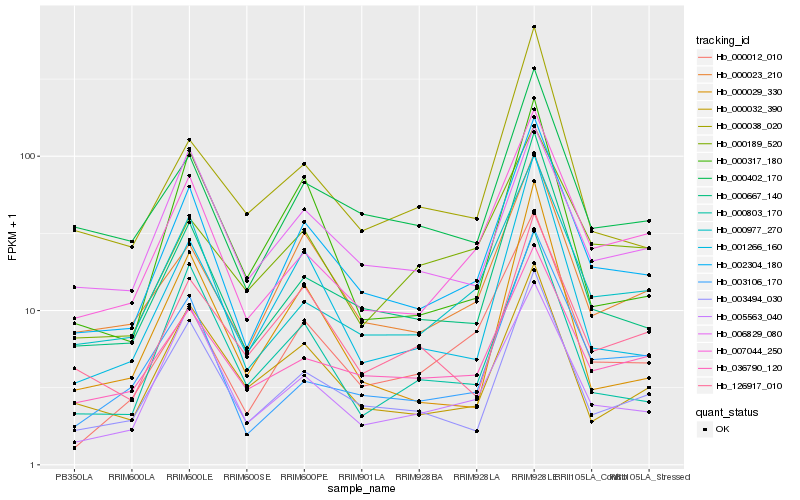
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002304_180 |
0.0 |
- |
- |
PREDICTED: magnesium-chelatase subunit ChlI, chloroplastic [Jatropha curcas] |
| 2 |
Hb_003494_030 |
0.0957892316 |
- |
- |
PREDICTED: protease Do-like 8, chloroplastic [Jatropha curcas] |
| 3 |
Hb_000667_140 |
0.0996025548 |
- |
- |
PREDICTED: uridine kinase-like protein 3 isoform X1 [Jatropha curcas] |
| 4 |
Hb_000977_270 |
0.1028643596 |
- |
- |
PREDICTED: phosphoglycolate phosphatase 1B, chloroplastic [Jatropha curcas] |
| 5 |
Hb_000023_210 |
0.1037851382 |
- |
- |
PREDICTED: protein TIC 55, chloroplastic [Jatropha curcas] |
| 6 |
Hb_007044_250 |
0.1042727035 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_000402_170 |
0.1140588609 |
- |
- |
PREDICTED: glutamate--glyoxylate aminotransferase 2 [Jatropha curcas] |
| 8 |
Hb_000012_010 |
0.1146836611 |
- |
- |
hypothetical protein JCGZ_07709 [Jatropha curcas] |
| 9 |
Hb_001266_160 |
0.118225556 |
- |
- |
PREDICTED: pheophytinase, chloroplastic [Jatropha curcas] |
| 10 |
Hb_003106_170 |
0.1232274451 |
- |
- |
Endonuclease III, putative [Ricinus communis] |
| 11 |
Hb_036790_120 |
0.1235383839 |
- |
- |
PREDICTED: uncharacterized protein LOC105645122 isoform X1 [Jatropha curcas] |
| 12 |
Hb_005563_040 |
0.1294941922 |
- |
- |
PREDICTED: uncharacterized protein LOC105630105 isoform X1 [Jatropha curcas] |
| 13 |
Hb_000032_390 |
0.1366573526 |
- |
- |
PREDICTED: uncharacterized protein LOC105645663 [Jatropha curcas] |
| 14 |
Hb_126917_010 |
0.137573088 |
- |
- |
PREDICTED: uncharacterized protein LOC105646333 [Jatropha curcas] |
| 15 |
Hb_000803_170 |
0.1396435793 |
- |
- |
PREDICTED: uncharacterized protein LOC105648312 [Jatropha curcas] |
| 16 |
Hb_000317_180 |
0.1439846714 |
- |
- |
PREDICTED: protein THYLAKOID FORMATION1, chloroplastic [Jatropha curcas] |
| 17 |
Hb_006829_080 |
0.144485952 |
- |
- |
PREDICTED: protein LHCP TRANSLOCATION DEFECT [Jatropha curcas] |
| 18 |
Hb_000029_330 |
0.1451305217 |
- |
- |
PREDICTED: protein TIC 62, chloroplastic [Jatropha curcas] |
| 19 |
Hb_000189_520 |
0.14554003 |
- |
- |
PREDICTED: uncharacterized protein LOC105647658 [Jatropha curcas] |
| 20 |
Hb_000038_020 |
0.1478211665 |
- |
- |
PREDICTED: ATP-dependent zinc metalloprotease FTSH 2, chloroplastic [Jatropha curcas] |