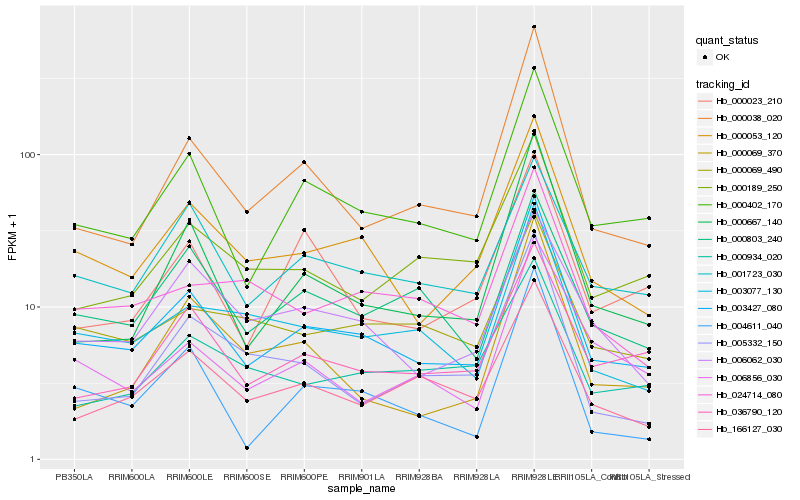
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003427_080 |
0.0 |
- |
- |
PREDICTED: probable alanine--tRNA ligase, chloroplastic [Jatropha curcas] |
| 2 |
Hb_000402_170 |
0.0802681259 |
- |
- |
PREDICTED: glutamate--glyoxylate aminotransferase 2 [Jatropha curcas] |
| 3 |
Hb_000053_120 |
0.0869974238 |
- |
- |
PREDICTED: protein CHUP1, chloroplastic [Jatropha curcas] |
| 4 |
Hb_000038_020 |
0.1070857614 |
- |
- |
PREDICTED: ATP-dependent zinc metalloprotease FTSH 2, chloroplastic [Jatropha curcas] |
| 5 |
Hb_000667_140 |
0.1083918546 |
- |
- |
PREDICTED: uridine kinase-like protein 3 isoform X1 [Jatropha curcas] |
| 6 |
Hb_003077_130 |
0.1250154449 |
- |
- |
hypothetical protein JCGZ_22476 [Jatropha curcas] |
| 7 |
Hb_166127_030 |
0.1285778335 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 8 |
Hb_036790_120 |
0.1300034302 |
- |
- |
PREDICTED: uncharacterized protein LOC105645122 isoform X1 [Jatropha curcas] |
| 9 |
Hb_004611_040 |
0.1304789409 |
- |
- |
PREDICTED: RNA polymerase sigma factor sigD, chloroplastic isoform X1 [Jatropha curcas] |
| 10 |
Hb_000189_250 |
0.1365432286 |
- |
- |
PREDICTED: probable ribose-5-phosphate isomerase 3, chloroplastic [Jatropha curcas] |
| 11 |
Hb_024714_080 |
0.1388066002 |
- |
- |
PREDICTED: translocase of chloroplast 159, chloroplastic [Jatropha curcas] |
| 12 |
Hb_006856_030 |
0.1389724637 |
- |
- |
Uridine kinase-like protein 4 [Glycine soja] |
| 13 |
Hb_001723_030 |
0.1391208052 |
- |
- |
PREDICTED: 30S ribosomal protein S1, chloroplastic [Jatropha curcas] |
| 14 |
Hb_000803_240 |
0.1418156365 |
- |
- |
hypothetical protein CICLE_v100143612mg, partial [Citrus clementina] |
| 15 |
Hb_000934_020 |
0.1431980982 |
- |
- |
PREDICTED: uncharacterized protein LOC105629153 [Jatropha curcas] |
| 16 |
Hb_000023_210 |
0.1435471904 |
- |
- |
PREDICTED: protein TIC 55, chloroplastic [Jatropha curcas] |
| 17 |
Hb_000069_490 |
0.1442472914 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_006062_030 |
0.1447325453 |
- |
- |
PREDICTED: translation initiation factor IF-2, chloroplastic [Jatropha curcas] |
| 19 |
Hb_000069_370 |
0.1453907956 |
- |
- |
- |
| 20 |
Hb_005332_150 |
0.145561139 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g46610 [Jatropha curcas] |