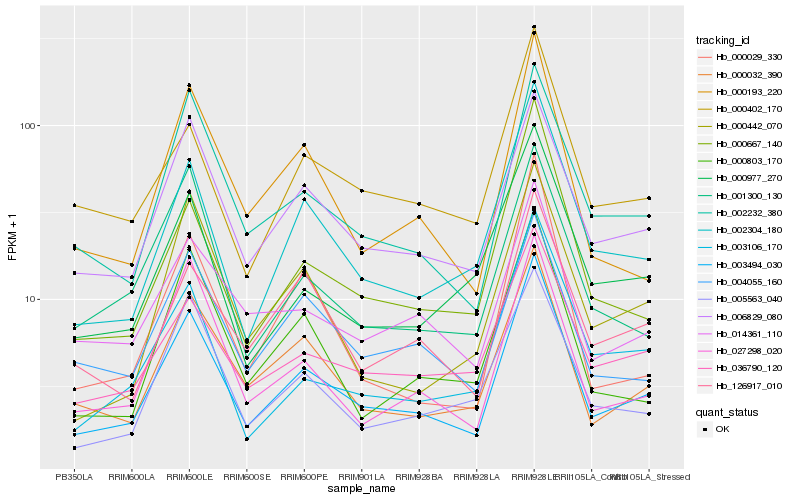
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003494_030 |
0.0 |
- |
- |
PREDICTED: protease Do-like 8, chloroplastic [Jatropha curcas] |
| 2 |
Hb_002304_180 |
0.0957892316 |
- |
- |
PREDICTED: magnesium-chelatase subunit ChlI, chloroplastic [Jatropha curcas] |
| 3 |
Hb_000032_390 |
0.1020908947 |
- |
- |
PREDICTED: uncharacterized protein LOC105645663 [Jatropha curcas] |
| 4 |
Hb_002232_380 |
0.1090421943 |
- |
- |
malate dehydrogenase, putative [Ricinus communis] |
| 5 |
Hb_006829_080 |
0.1109685725 |
- |
- |
PREDICTED: protein LHCP TRANSLOCATION DEFECT [Jatropha curcas] |
| 6 |
Hb_000193_220 |
0.1117458892 |
- |
- |
PREDICTED: ribulose-phosphate 3-epimerase, chloroplastic [Jatropha curcas] |
| 7 |
Hb_000667_140 |
0.1141401822 |
- |
- |
PREDICTED: uridine kinase-like protein 3 isoform X1 [Jatropha curcas] |
| 8 |
Hb_027298_020 |
0.1191669694 |
- |
- |
hypothetical protein POPTR_0005s05760g [Populus trichocarpa] |
| 9 |
Hb_036790_120 |
0.1205608422 |
- |
- |
PREDICTED: uncharacterized protein LOC105645122 isoform X1 [Jatropha curcas] |
| 10 |
Hb_000803_170 |
0.121027152 |
- |
- |
PREDICTED: uncharacterized protein LOC105648312 [Jatropha curcas] |
| 11 |
Hb_005563_040 |
0.1219668798 |
- |
- |
PREDICTED: uncharacterized protein LOC105630105 isoform X1 [Jatropha curcas] |
| 12 |
Hb_004055_160 |
0.1242506186 |
- |
- |
PREDICTED: glyoxylate/succinic semialdehyde reductase 2, chloroplastic [Jatropha curcas] |
| 13 |
Hb_000402_170 |
0.1243999556 |
- |
- |
PREDICTED: glutamate--glyoxylate aminotransferase 2 [Jatropha curcas] |
| 14 |
Hb_126917_010 |
0.1257584446 |
- |
- |
PREDICTED: uncharacterized protein LOC105646333 [Jatropha curcas] |
| 15 |
Hb_000029_330 |
0.1291010951 |
- |
- |
PREDICTED: protein TIC 62, chloroplastic [Jatropha curcas] |
| 16 |
Hb_000977_270 |
0.1292858519 |
- |
- |
PREDICTED: phosphoglycolate phosphatase 1B, chloroplastic [Jatropha curcas] |
| 17 |
Hb_003106_170 |
0.130208629 |
- |
- |
Endonuclease III, putative [Ricinus communis] |
| 18 |
Hb_014361_110 |
0.1313687714 |
- |
- |
RNA binding protein, putative [Ricinus communis] |
| 19 |
Hb_000442_070 |
0.1318168697 |
- |
- |
PREDICTED: uncharacterized protein LOC105645538 [Jatropha curcas] |
| 20 |
Hb_001300_130 |
0.1320001562 |
- |
- |
PREDICTED: protein TRANSPORT INHIBITOR RESPONSE 1 [Jatropha curcas] |