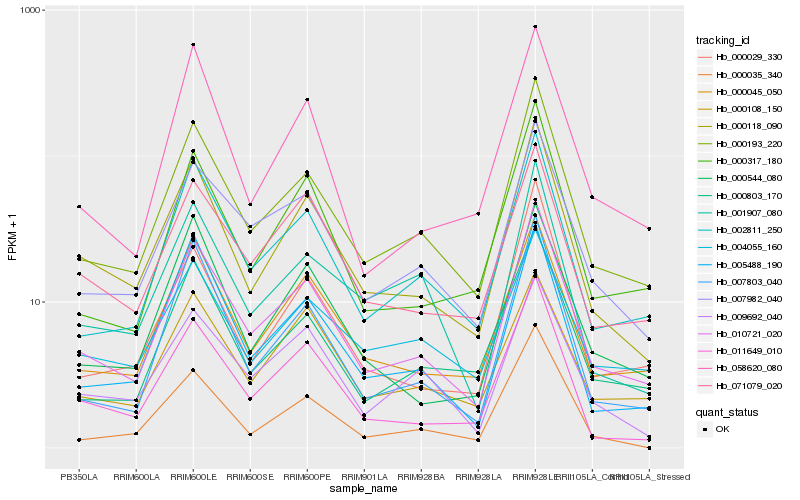
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000118_090 |
0.0 |
- |
- |
Glutamyl-tRNA reductase 1, chloroplast precursor, putative [Ricinus communis] |
| 2 |
Hb_010721_020 |
0.0788316902 |
- |
- |
PREDICTED: photosynthetic NDH subunit of lumenal location 4, chloroplastic [Jatropha curcas] |
| 3 |
Hb_011649_010 |
0.0814642719 |
transcription factor |
TF Family: Pseudo ARR-B |
sensory transduction histidine kinase, putative [Ricinus communis] |
| 4 |
Hb_000193_220 |
0.0890896497 |
- |
- |
PREDICTED: ribulose-phosphate 3-epimerase, chloroplastic [Jatropha curcas] |
| 5 |
Hb_000045_050 |
0.107262714 |
- |
- |
PREDICTED: probable zinc metalloprotease EGY2, chloroplastic [Jatropha curcas] |
| 6 |
Hb_071079_020 |
0.1120626328 |
- |
- |
RNA polymerase sigma factor rpoD, putative [Ricinus communis] |
| 7 |
Hb_000544_080 |
0.1237523378 |
- |
- |
PREDICTED: uncharacterized protein LOC105631062 isoform X3 [Jatropha curcas] |
| 8 |
Hb_009692_040 |
0.1252140138 |
- |
- |
PREDICTED: peptide chain release factor PrfB3, chloroplastic isoform X1 [Jatropha curcas] |
| 9 |
Hb_007982_040 |
0.1257528458 |
- |
- |
PREDICTED: uncharacterized aarF domain-containing protein kinase At1g79600, chloroplastic [Jatropha curcas] |
| 10 |
Hb_005488_190 |
0.1263559814 |
- |
- |
RNA polymerase sigma factor rpoD, putative [Ricinus communis] |
| 11 |
Hb_002811_250 |
0.1268075849 |
- |
- |
PREDICTED: probable ribose-5-phosphate isomerase 3, chloroplastic [Jatropha curcas] |
| 12 |
Hb_001907_080 |
0.1282948149 |
- |
- |
PREDICTED: uncharacterized protein LOC105628749 [Jatropha curcas] |
| 13 |
Hb_000317_180 |
0.12936678 |
- |
- |
PREDICTED: protein THYLAKOID FORMATION1, chloroplastic [Jatropha curcas] |
| 14 |
Hb_000035_340 |
0.1297499318 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_000029_330 |
0.1307836539 |
- |
- |
PREDICTED: protein TIC 62, chloroplastic [Jatropha curcas] |
| 16 |
Hb_004055_160 |
0.1329741192 |
- |
- |
PREDICTED: glyoxylate/succinic semialdehyde reductase 2, chloroplastic [Jatropha curcas] |
| 17 |
Hb_007803_040 |
0.133432895 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase CYP37, chloroplastic [Jatropha curcas] |
| 18 |
Hb_058620_080 |
0.1337515504 |
- |
- |
PREDICTED: phosphoglycerate kinase, chloroplastic [Jatropha curcas] |
| 19 |
Hb_000803_170 |
0.1348309209 |
- |
- |
PREDICTED: uncharacterized protein LOC105648312 [Jatropha curcas] |
| 20 |
Hb_000108_150 |
0.1352133716 |
- |
- |
alpha/beta hydrolase, putative [Ricinus communis] |