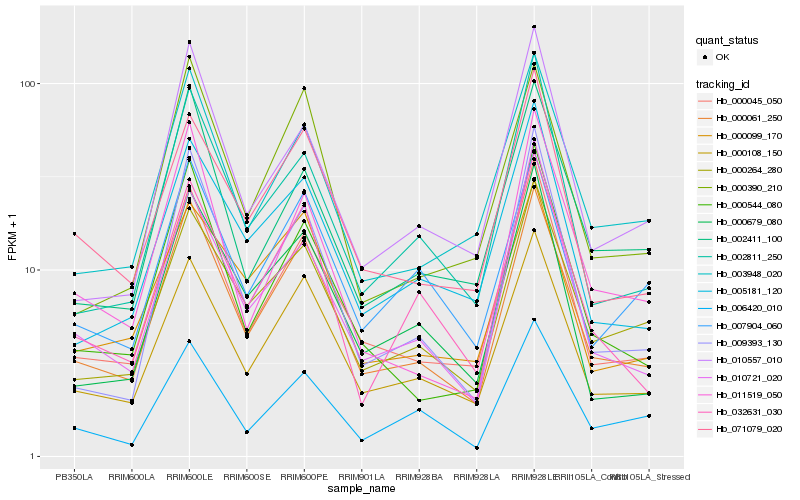
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000061_250 |
0.0 |
- |
- |
hypothetical protein JCGZ_11665 [Jatropha curcas] |
| 2 |
Hb_000108_150 |
0.0595748299 |
- |
- |
alpha/beta hydrolase, putative [Ricinus communis] |
| 3 |
Hb_000045_050 |
0.0683445589 |
- |
- |
PREDICTED: probable zinc metalloprotease EGY2, chloroplastic [Jatropha curcas] |
| 4 |
Hb_000544_080 |
0.0906753356 |
- |
- |
PREDICTED: uncharacterized protein LOC105631062 isoform X3 [Jatropha curcas] |
| 5 |
Hb_000264_280 |
0.0911540255 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RMA1H1-like [Jatropha curcas] |
| 6 |
Hb_003948_020 |
0.091770205 |
- |
- |
PREDICTED: transmembrane protein 41B [Jatropha curcas] |
| 7 |
Hb_000390_210 |
0.0981499877 |
- |
- |
cinnamoyl-CoA reductase, putative [Ricinus communis] |
| 8 |
Hb_071079_020 |
0.0996896494 |
- |
- |
RNA polymerase sigma factor rpoD, putative [Ricinus communis] |
| 9 |
Hb_032631_030 |
0.1007745129 |
- |
- |
phosphatidic acid phosphatase, putative [Ricinus communis] |
| 10 |
Hb_002411_100 |
0.1051300094 |
- |
- |
PREDICTED: uncharacterized protein LOC105631266 [Jatropha curcas] |
| 11 |
Hb_005181_120 |
0.1055890906 |
- |
- |
PREDICTED: psbB mRNA maturation factor Mbb1, chloroplastic isoform X1 [Jatropha curcas] |
| 12 |
Hb_000679_080 |
0.1062482406 |
- |
- |
PREDICTED: lactation elevated protein 1 isoform X1 [Jatropha curcas] |
| 13 |
Hb_009393_130 |
0.1070716851 |
- |
- |
PREDICTED: uncharacterized protein LOC105638053 isoform X4 [Jatropha curcas] |
| 14 |
Hb_007904_060 |
0.1073085441 |
- |
- |
PREDICTED: very-long-chain 3-oxoacyl-CoA reductase 1 [Jatropha curcas] |
| 15 |
Hb_010557_010 |
0.1085847679 |
- |
- |
PREDICTED: 50S ribosomal protein L3, chloroplastic [Jatropha curcas] |
| 16 |
Hb_002811_250 |
0.109787976 |
- |
- |
PREDICTED: probable ribose-5-phosphate isomerase 3, chloroplastic [Jatropha curcas] |
| 17 |
Hb_011519_050 |
0.1099965462 |
- |
- |
hypothetical protein POPTR_0015s04810g [Populus trichocarpa] |
| 18 |
Hb_000099_170 |
0.1104053498 |
- |
- |
PREDICTED: uncharacterized protein LOC105645733 [Jatropha curcas] |
| 19 |
Hb_010721_020 |
0.1105868626 |
- |
- |
PREDICTED: photosynthetic NDH subunit of lumenal location 4, chloroplastic [Jatropha curcas] |
| 20 |
Hb_006420_010 |
0.1114136125 |
- |
- |
PREDICTED: uncharacterized protein LOC105649815 [Jatropha curcas] |