| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
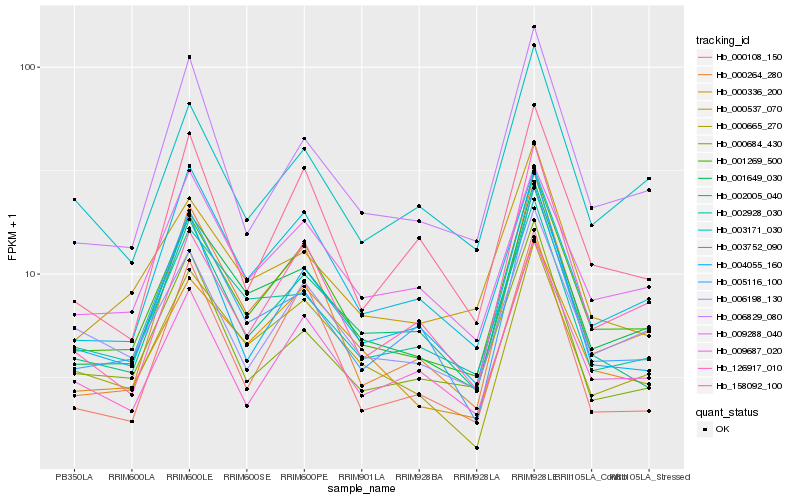
Hb_001269_500 |
0.0 |
- |
- |
PREDICTED: protein LOW PSII ACCUMULATION 3, chloroplastic-like isoform X2 [Populus euphratica] |
| 2 |
Hb_001649_030 |
0.0657737463 |
- |
- |
PREDICTED: peptide deformylase 1B, chloroplastic [Jatropha curcas] |
| 3 |
Hb_000336_200 |
0.07856038 |
- |
- |
PREDICTED: UPF0481 protein At3g47200-like [Jatropha curcas] |
| 4 |
Hb_009687_020 |
0.0854621748 |
- |
- |
hypothetical protein JCGZ_10323 [Jatropha curcas] |
| 5 |
Hb_000665_270 |
0.0884565176 |
- |
- |
PREDICTED: adenine phosphoribosyltransferase 5 [Jatropha curcas] |
| 6 |
Hb_000264_280 |
0.0920778291 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RMA1H1-like [Jatropha curcas] |
| 7 |
Hb_006198_130 |
0.0925985955 |
- |
- |
PREDICTED: uncharacterized protein LOC105644406 [Jatropha curcas] |
| 8 |
Hb_002005_040 |
0.0936680998 |
- |
- |
PREDICTED: carboxyl-terminal-processing peptidase 1, chloroplastic isoform X2 [Jatropha curcas] |
| 9 |
Hb_006829_080 |
0.0938051215 |
- |
- |
PREDICTED: protein LHCP TRANSLOCATION DEFECT [Jatropha curcas] |
| 10 |
Hb_004055_160 |
0.0995634786 |
- |
- |
PREDICTED: glyoxylate/succinic semialdehyde reductase 2, chloroplastic [Jatropha curcas] |
| 11 |
Hb_009288_040 |
0.1004482801 |
- |
- |
PREDICTED: uncharacterized protein LOC105648677 [Jatropha curcas] |
| 12 |
Hb_003752_090 |
0.100794441 |
- |
- |
chitinase, putative [Ricinus communis] |
| 13 |
Hb_002928_030 |
0.1010957148 |
- |
- |
hypothetical protein CICLE_v10001788mg [Citrus clementina] |
| 14 |
Hb_003171_030 |
0.1014747554 |
- |
- |
PREDICTED: grpE protein homolog, mitochondrial [Jatropha curcas] |
| 15 |
Hb_000537_070 |
0.102373224 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g25630 [Jatropha curcas] |
| 16 |
Hb_158092_100 |
0.1044810248 |
- |
- |
PREDICTED: uncharacterized protein LOC105647301 [Jatropha curcas] |
| 17 |
Hb_000684_430 |
0.1102163009 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g02830, chloroplastic [Jatropha curcas] |
| 18 |
Hb_005116_100 |
0.1107426793 |
- |
- |
PREDICTED: uncharacterized protein LOC105649999 [Jatropha curcas] |
| 19 |
Hb_000108_150 |
0.1109313447 |
- |
- |
alpha/beta hydrolase, putative [Ricinus communis] |
| 20 |
Hb_126917_010 |
0.1143251087 |
- |
- |
PREDICTED: uncharacterized protein LOC105646333 [Jatropha curcas] |