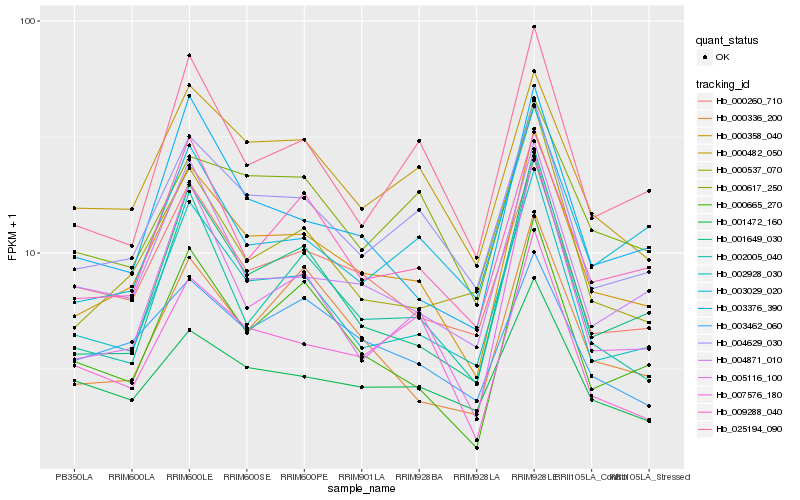
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000358_040 |
0.0 |
- |
- |
PREDICTED: probable plastid-lipid-associated protein 4, chloroplastic [Jatropha curcas] |
| 2 |
Hb_002928_030 |
0.0825474592 |
- |
- |
hypothetical protein CICLE_v10001788mg [Citrus clementina] |
| 3 |
Hb_004629_030 |
0.0830280694 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 4 |
Hb_001649_030 |
0.089912513 |
- |
- |
PREDICTED: peptide deformylase 1B, chloroplastic [Jatropha curcas] |
| 5 |
Hb_000260_710 |
0.092005405 |
- |
- |
PREDICTED: uncharacterized protein LOC105649017 isoform X1 [Jatropha curcas] |
| 6 |
Hb_000482_050 |
0.0947880563 |
- |
- |
PREDICTED: UDP-sulfoquinovose synthase, chloroplastic [Jatropha curcas] |
| 7 |
Hb_003376_390 |
0.0970006606 |
- |
- |
PREDICTED: 29 kDa ribonucleoprotein A, chloroplastic [Jatropha curcas] |
| 8 |
Hb_009288_040 |
0.0973065188 |
- |
- |
PREDICTED: uncharacterized protein LOC105648677 [Jatropha curcas] |
| 9 |
Hb_000665_270 |
0.0985090808 |
- |
- |
PREDICTED: adenine phosphoribosyltransferase 5 [Jatropha curcas] |
| 10 |
Hb_004871_010 |
0.1007074713 |
- |
- |
RNA polymerase sigma factor rpoD1, putative [Ricinus communis] |
| 11 |
Hb_025194_090 |
0.104458132 |
- |
- |
coproporphyrinogen III oxidase, putative [Ricinus communis] |
| 12 |
Hb_005116_100 |
0.1104079699 |
- |
- |
PREDICTED: uncharacterized protein LOC105649999 [Jatropha curcas] |
| 13 |
Hb_002005_040 |
0.1107157562 |
- |
- |
PREDICTED: carboxyl-terminal-processing peptidase 1, chloroplastic isoform X2 [Jatropha curcas] |
| 14 |
Hb_000336_200 |
0.1114564232 |
- |
- |
PREDICTED: UPF0481 protein At3g47200-like [Jatropha curcas] |
| 15 |
Hb_000537_070 |
0.1137001573 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g25630 [Jatropha curcas] |
| 16 |
Hb_000617_250 |
0.1137249817 |
- |
- |
PREDICTED: ATP-dependent Clp protease proteolytic subunit-related protein 3, chloroplastic [Jatropha curcas] |
| 17 |
Hb_001472_160 |
0.1140183342 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase kinase YODA [Jatropha curcas] |
| 18 |
Hb_007576_180 |
0.114368628 |
- |
- |
PREDICTED: probable protein phosphatase 2C 62 isoform X5 [Jatropha curcas] |
| 19 |
Hb_003029_020 |
0.1143781199 |
- |
- |
ATP-dependent Clp protease proteolytic subunit, putative [Ricinus communis] |
| 20 |
Hb_003462_060 |
0.1145837866 |
- |
- |
PREDICTED: ABC transporter G family member 24-like [Jatropha curcas] |