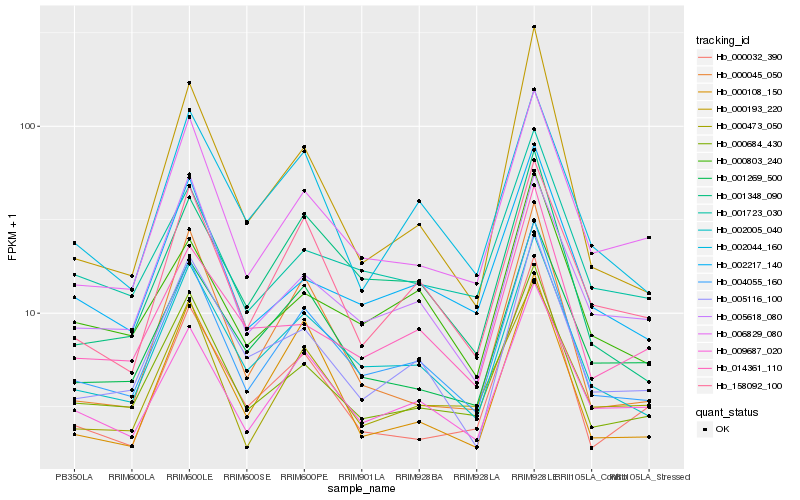
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004055_160 |
0.0 |
- |
- |
PREDICTED: glyoxylate/succinic semialdehyde reductase 2, chloroplastic [Jatropha curcas] |
| 2 |
Hb_002005_040 |
0.056136248 |
- |
- |
PREDICTED: carboxyl-terminal-processing peptidase 1, chloroplastic isoform X2 [Jatropha curcas] |
| 3 |
Hb_000684_430 |
0.0744007651 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g02830, chloroplastic [Jatropha curcas] |
| 4 |
Hb_009687_020 |
0.083242132 |
- |
- |
hypothetical protein JCGZ_10323 [Jatropha curcas] |
| 5 |
Hb_006829_080 |
0.0839176699 |
- |
- |
PREDICTED: protein LHCP TRANSLOCATION DEFECT [Jatropha curcas] |
| 6 |
Hb_005116_100 |
0.0903074417 |
- |
- |
PREDICTED: uncharacterized protein LOC105649999 [Jatropha curcas] |
| 7 |
Hb_001348_090 |
0.0913383851 |
- |
- |
PREDICTED: long chain acyl-CoA synthetase 9, chloroplastic [Jatropha curcas] |
| 8 |
Hb_002217_140 |
0.0938284546 |
- |
- |
PREDICTED: 30S ribosomal protein S1, chloroplastic [Nelumbo nucifera] |
| 9 |
Hb_158092_100 |
0.0944245164 |
- |
- |
PREDICTED: uncharacterized protein LOC105647301 [Jatropha curcas] |
| 10 |
Hb_001269_500 |
0.0995634786 |
- |
- |
PREDICTED: protein LOW PSII ACCUMULATION 3, chloroplastic-like isoform X2 [Populus euphratica] |
| 11 |
Hb_002044_160 |
0.1015692192 |
- |
- |
PREDICTED: glyoxylate/succinic semialdehyde reductase 1 [Jatropha curcas] |
| 12 |
Hb_000473_050 |
0.1031464959 |
- |
- |
chromatin regulatory protein sir2, putative [Ricinus communis] |
| 13 |
Hb_000803_240 |
0.103732039 |
- |
- |
hypothetical protein CICLE_v100143612mg, partial [Citrus clementina] |
| 14 |
Hb_001723_030 |
0.1055279067 |
- |
- |
PREDICTED: 30S ribosomal protein S1, chloroplastic [Jatropha curcas] |
| 15 |
Hb_000108_150 |
0.1057767572 |
- |
- |
alpha/beta hydrolase, putative [Ricinus communis] |
| 16 |
Hb_000032_390 |
0.1080924338 |
- |
- |
PREDICTED: uncharacterized protein LOC105645663 [Jatropha curcas] |
| 17 |
Hb_014361_110 |
0.1091699069 |
- |
- |
RNA binding protein, putative [Ricinus communis] |
| 18 |
Hb_005618_080 |
0.1092619828 |
- |
- |
PREDICTED: protoporphyrinogen oxidase 1, chloroplastic [Jatropha curcas] |
| 19 |
Hb_000045_050 |
0.1118685896 |
- |
- |
PREDICTED: probable zinc metalloprotease EGY2, chloroplastic [Jatropha curcas] |
| 20 |
Hb_000193_220 |
0.1125232436 |
- |
- |
PREDICTED: ribulose-phosphate 3-epimerase, chloroplastic [Jatropha curcas] |