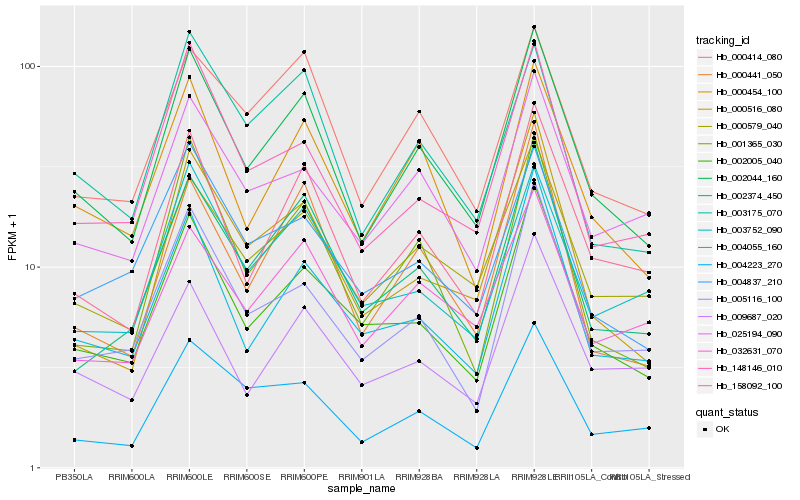
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002044_160 |
0.0 |
- |
- |
PREDICTED: glyoxylate/succinic semialdehyde reductase 1 [Jatropha curcas] |
| 2 |
Hb_158092_100 |
0.0776342982 |
- |
- |
PREDICTED: uncharacterized protein LOC105647301 [Jatropha curcas] |
| 3 |
Hb_000454_100 |
0.0810582032 |
- |
- |
aldo-keto reductase, putative [Ricinus communis] |
| 4 |
Hb_002005_040 |
0.0876688831 |
- |
- |
PREDICTED: carboxyl-terminal-processing peptidase 1, chloroplastic isoform X2 [Jatropha curcas] |
| 5 |
Hb_000579_040 |
0.0891009779 |
- |
- |
PREDICTED: putative GTP-binding protein 6 [Jatropha curcas] |
| 6 |
Hb_002374_450 |
0.097428963 |
- |
- |
PREDICTED: chlorophyllide a oxygenase, chloroplastic [Jatropha curcas] |
| 7 |
Hb_004055_160 |
0.1015692192 |
- |
- |
PREDICTED: glyoxylate/succinic semialdehyde reductase 2, chloroplastic [Jatropha curcas] |
| 8 |
Hb_000414_080 |
0.1021436228 |
- |
- |
PREDICTED: triosephosphate isomerase, chloroplastic [Jatropha curcas] |
| 9 |
Hb_003175_070 |
0.1042295964 |
- |
- |
pyrophosphate-dependent phosphofructokinase, partial [Hevea brasiliensis] |
| 10 |
Hb_005116_100 |
0.1053008161 |
- |
- |
PREDICTED: uncharacterized protein LOC105649999 [Jatropha curcas] |
| 11 |
Hb_004223_270 |
0.1059956481 |
- |
- |
PREDICTED: uncharacterized protein LOC105635371 isoform X1 [Jatropha curcas] |
| 12 |
Hb_001365_030 |
0.1061069608 |
- |
- |
PREDICTED: uncharacterized protein LOC102609547 [Citrus sinensis] |
| 13 |
Hb_000441_050 |
0.1068427906 |
- |
- |
PREDICTED: 187-kDa microtubule-associated protein AIR9 [Jatropha curcas] |
| 14 |
Hb_025194_090 |
0.1068704772 |
- |
- |
coproporphyrinogen III oxidase, putative [Ricinus communis] |
| 15 |
Hb_148146_010 |
0.1079261078 |
- |
- |
coproporphyrinogen III oxidase [Ziziphus jujuba] |
| 16 |
Hb_000516_080 |
0.1084679697 |
- |
- |
hypothetical protein POPTR_0010s24810g [Populus trichocarpa] |
| 17 |
Hb_003752_090 |
0.1087400144 |
- |
- |
chitinase, putative [Ricinus communis] |
| 18 |
Hb_032631_070 |
0.1087748946 |
- |
- |
PREDICTED: lisH domain and HEAT repeat-containing protein KIAA1468 homolog isoform X1 [Jatropha curcas] |
| 19 |
Hb_004837_210 |
0.1119374358 |
- |
- |
PREDICTED: calcium-dependent protein kinase 8-like [Jatropha curcas] |
| 20 |
Hb_009687_020 |
0.1131802879 |
- |
- |
hypothetical protein JCGZ_10323 [Jatropha curcas] |