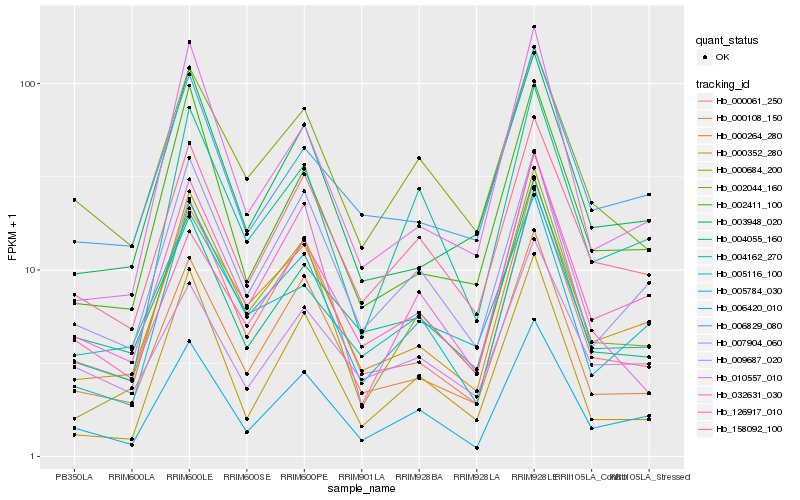
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006420_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105649815 [Jatropha curcas] |
| 2 |
Hb_158092_100 |
0.0834902613 |
- |
- |
PREDICTED: uncharacterized protein LOC105647301 [Jatropha curcas] |
| 3 |
Hb_007904_060 |
0.0908413739 |
- |
- |
PREDICTED: very-long-chain 3-oxoacyl-CoA reductase 1 [Jatropha curcas] |
| 4 |
Hb_000264_280 |
0.1068655649 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RMA1H1-like [Jatropha curcas] |
| 5 |
Hb_000108_150 |
0.1087088807 |
- |
- |
alpha/beta hydrolase, putative [Ricinus communis] |
| 6 |
Hb_000061_250 |
0.1114136125 |
- |
- |
hypothetical protein JCGZ_11665 [Jatropha curcas] |
| 7 |
Hb_004162_270 |
0.1136657904 |
- |
- |
PREDICTED: superoxide dismutase [Cu-Zn], chloroplastic isoform X1 [Jatropha curcas] |
| 8 |
Hb_004055_160 |
0.1212155927 |
- |
- |
PREDICTED: glyoxylate/succinic semialdehyde reductase 2, chloroplastic [Jatropha curcas] |
| 9 |
Hb_000684_200 |
0.1218092585 |
- |
- |
PREDICTED: protein IQ-DOMAIN 1-like [Jatropha curcas] |
| 10 |
Hb_002411_100 |
0.122089441 |
- |
- |
PREDICTED: uncharacterized protein LOC105631266 [Jatropha curcas] |
| 11 |
Hb_005784_030 |
0.1234616718 |
- |
- |
PREDICTED: uncharacterized protein LOC105645162 isoform X2 [Jatropha curcas] |
| 12 |
Hb_126917_010 |
0.1257803577 |
- |
- |
PREDICTED: uncharacterized protein LOC105646333 [Jatropha curcas] |
| 13 |
Hb_009687_020 |
0.1264139472 |
- |
- |
hypothetical protein JCGZ_10323 [Jatropha curcas] |
| 14 |
Hb_000352_280 |
0.1277172913 |
- |
- |
PREDICTED: acetyl-coenzyme A synthetase, chloroplastic/glyoxysomal isoform X2 [Jatropha curcas] |
| 15 |
Hb_005116_100 |
0.1278728903 |
- |
- |
PREDICTED: uncharacterized protein LOC105649999 [Jatropha curcas] |
| 16 |
Hb_006829_080 |
0.1291673145 |
- |
- |
PREDICTED: protein LHCP TRANSLOCATION DEFECT [Jatropha curcas] |
| 17 |
Hb_002044_160 |
0.1298641541 |
- |
- |
PREDICTED: glyoxylate/succinic semialdehyde reductase 1 [Jatropha curcas] |
| 18 |
Hb_032631_030 |
0.1300943882 |
- |
- |
phosphatidic acid phosphatase, putative [Ricinus communis] |
| 19 |
Hb_003948_020 |
0.1306849429 |
- |
- |
PREDICTED: transmembrane protein 41B [Jatropha curcas] |
| 20 |
Hb_010557_010 |
0.1319208287 |
- |
- |
PREDICTED: 50S ribosomal protein L3, chloroplastic [Jatropha curcas] |