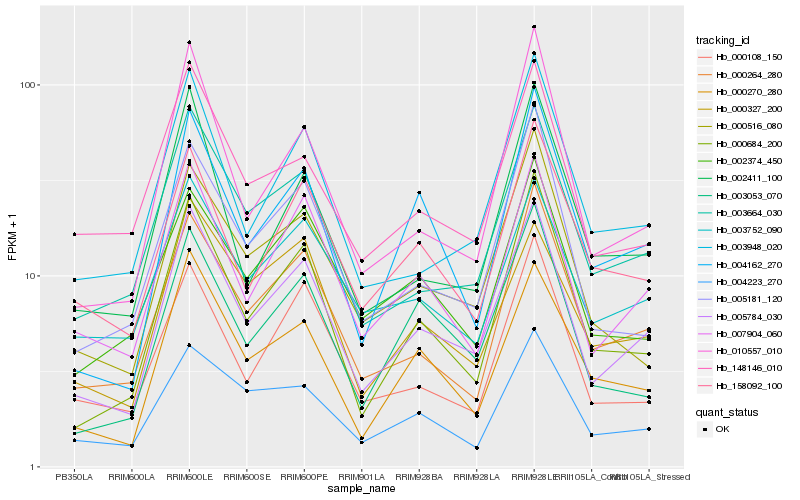
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005784_030 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105645162 isoform X2 [Jatropha curcas] |
| 2 |
Hb_007904_060 |
0.0811072227 |
- |
- |
PREDICTED: very-long-chain 3-oxoacyl-CoA reductase 1 [Jatropha curcas] |
| 3 |
Hb_003664_030 |
0.0912703427 |
- |
- |
PREDICTED: mitochondrial carnitine/acylcarnitine carrier-like protein [Jatropha curcas] |
| 4 |
Hb_000264_280 |
0.1004557528 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RMA1H1-like [Jatropha curcas] |
| 5 |
Hb_000684_200 |
0.1012132207 |
- |
- |
PREDICTED: protein IQ-DOMAIN 1-like [Jatropha curcas] |
| 6 |
Hb_000270_280 |
0.106323947 |
- |
- |
Rhicadhesin receptor precursor, putative [Ricinus communis] |
| 7 |
Hb_003948_020 |
0.1063311301 |
- |
- |
PREDICTED: transmembrane protein 41B [Jatropha curcas] |
| 8 |
Hb_004162_270 |
0.112083067 |
- |
- |
PREDICTED: superoxide dismutase [Cu-Zn], chloroplastic isoform X1 [Jatropha curcas] |
| 9 |
Hb_003752_090 |
0.1126973859 |
- |
- |
chitinase, putative [Ricinus communis] |
| 10 |
Hb_010557_010 |
0.1141692296 |
- |
- |
PREDICTED: 50S ribosomal protein L3, chloroplastic [Jatropha curcas] |
| 11 |
Hb_158092_100 |
0.1143811284 |
- |
- |
PREDICTED: uncharacterized protein LOC105647301 [Jatropha curcas] |
| 12 |
Hb_000327_200 |
0.114479249 |
- |
- |
glutathione-s-transferase omega, putative [Ricinus communis] |
| 13 |
Hb_003053_070 |
0.1148435532 |
- |
- |
PREDICTED: phosphoglucan phosphatase LSF1, chloroplastic [Jatropha curcas] |
| 14 |
Hb_004223_270 |
0.1155240018 |
- |
- |
PREDICTED: uncharacterized protein LOC105635371 isoform X1 [Jatropha curcas] |
| 15 |
Hb_005181_120 |
0.1164176699 |
- |
- |
PREDICTED: psbB mRNA maturation factor Mbb1, chloroplastic isoform X1 [Jatropha curcas] |
| 16 |
Hb_000516_080 |
0.1176131646 |
- |
- |
hypothetical protein POPTR_0010s24810g [Populus trichocarpa] |
| 17 |
Hb_148146_010 |
0.1180706274 |
- |
- |
coproporphyrinogen III oxidase [Ziziphus jujuba] |
| 18 |
Hb_000108_150 |
0.1188252018 |
- |
- |
alpha/beta hydrolase, putative [Ricinus communis] |
| 19 |
Hb_002411_100 |
0.1195702418 |
- |
- |
PREDICTED: uncharacterized protein LOC105631266 [Jatropha curcas] |
| 20 |
Hb_002374_450 |
0.1221341364 |
- |
- |
PREDICTED: chlorophyllide a oxygenase, chloroplastic [Jatropha curcas] |