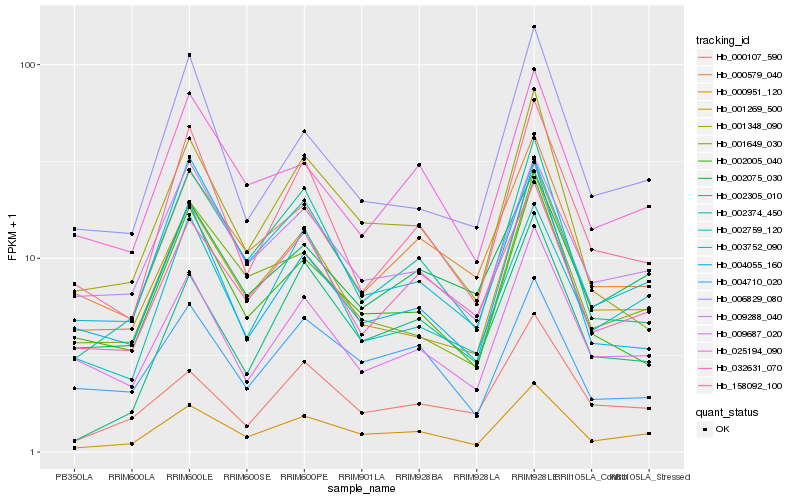
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000951_120 |
0.0 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC105636019 [Jatropha curcas] |
| 2 |
Hb_002374_450 |
0.0989885295 |
- |
- |
PREDICTED: chlorophyllide a oxygenase, chloroplastic [Jatropha curcas] |
| 3 |
Hb_032631_070 |
0.1044560238 |
- |
- |
PREDICTED: lisH domain and HEAT repeat-containing protein KIAA1468 homolog isoform X1 [Jatropha curcas] |
| 4 |
Hb_002075_030 |
0.1058602039 |
- |
- |
PREDICTED: glycerol-3-phosphate acyltransferase, chloroplastic [Jatropha curcas] |
| 5 |
Hb_158092_100 |
0.1134601721 |
- |
- |
PREDICTED: uncharacterized protein LOC105647301 [Jatropha curcas] |
| 6 |
Hb_001649_030 |
0.1149829538 |
- |
- |
PREDICTED: peptide deformylase 1B, chloroplastic [Jatropha curcas] |
| 7 |
Hb_003752_090 |
0.1151431404 |
- |
- |
chitinase, putative [Ricinus communis] |
| 8 |
Hb_001269_500 |
0.1160815727 |
- |
- |
PREDICTED: protein LOW PSII ACCUMULATION 3, chloroplastic-like isoform X2 [Populus euphratica] |
| 9 |
Hb_009687_020 |
0.1166621264 |
- |
- |
hypothetical protein JCGZ_10323 [Jatropha curcas] |
| 10 |
Hb_000579_040 |
0.1168643142 |
- |
- |
PREDICTED: putative GTP-binding protein 6 [Jatropha curcas] |
| 11 |
Hb_025194_090 |
0.117623353 |
- |
- |
coproporphyrinogen III oxidase, putative [Ricinus communis] |
| 12 |
Hb_002305_010 |
0.1176399883 |
- |
- |
S-adenosylmethionine-dependent methyltransferase, putative [Ricinus communis] |
| 13 |
Hb_002005_040 |
0.1183110973 |
- |
- |
PREDICTED: carboxyl-terminal-processing peptidase 1, chloroplastic isoform X2 [Jatropha curcas] |
| 14 |
Hb_000107_590 |
0.1193644915 |
- |
- |
PREDICTED: tricalbin-3 [Jatropha curcas] |
| 15 |
Hb_001348_090 |
0.1205741508 |
- |
- |
PREDICTED: long chain acyl-CoA synthetase 9, chloroplastic [Jatropha curcas] |
| 16 |
Hb_002759_120 |
0.1206711359 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_009288_040 |
0.1217647742 |
- |
- |
PREDICTED: uncharacterized protein LOC105648677 [Jatropha curcas] |
| 18 |
Hb_004710_020 |
0.1218059911 |
- |
- |
2-deoxyglucose-6-phosphate phosphatase, putative [Ricinus communis] |
| 19 |
Hb_006829_080 |
0.1218217187 |
- |
- |
PREDICTED: protein LHCP TRANSLOCATION DEFECT [Jatropha curcas] |
| 20 |
Hb_004055_160 |
0.1242769247 |
- |
- |
PREDICTED: glyoxylate/succinic semialdehyde reductase 2, chloroplastic [Jatropha curcas] |