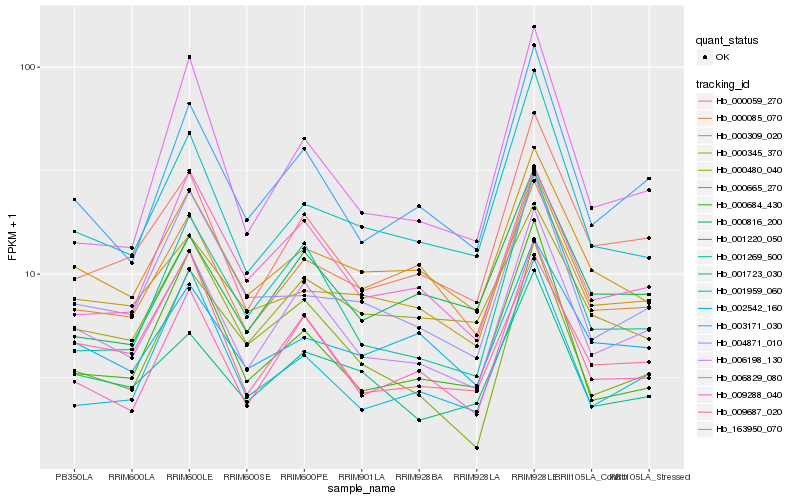
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006198_130 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105644406 [Jatropha curcas] |
| 2 |
Hb_003171_030 |
0.0706499773 |
- |
- |
PREDICTED: grpE protein homolog, mitochondrial [Jatropha curcas] |
| 3 |
Hb_009687_020 |
0.0723762354 |
- |
- |
hypothetical protein JCGZ_10323 [Jatropha curcas] |
| 4 |
Hb_000059_270 |
0.0824857262 |
- |
- |
superoxide dismutase [Fe], chloroplastic [Jatropha curcas] |
| 5 |
Hb_001269_500 |
0.0925985955 |
- |
- |
PREDICTED: protein LOW PSII ACCUMULATION 3, chloroplastic-like isoform X2 [Populus euphratica] |
| 6 |
Hb_163950_070 |
0.0982776971 |
- |
- |
arginine/serine-rich splicing factor, putative [Ricinus communis] |
| 7 |
Hb_001220_050 |
0.1020385284 |
- |
- |
DNA-damage-inducible protein f, putative [Ricinus communis] |
| 8 |
Hb_001723_030 |
0.1032413122 |
- |
- |
PREDICTED: 30S ribosomal protein S1, chloroplastic [Jatropha curcas] |
| 9 |
Hb_000309_020 |
0.1034115146 |
- |
- |
PREDICTED: uncharacterized protein LOC105641764 [Jatropha curcas] |
| 10 |
Hb_009288_040 |
0.1040334404 |
- |
- |
PREDICTED: uncharacterized protein LOC105648677 [Jatropha curcas] |
| 11 |
Hb_000665_270 |
0.11199788 |
- |
- |
PREDICTED: adenine phosphoribosyltransferase 5 [Jatropha curcas] |
| 12 |
Hb_000345_370 |
0.1131732314 |
- |
- |
PREDICTED: probable Xaa-Pro aminopeptidase P [Jatropha curcas] |
| 13 |
Hb_000480_040 |
0.1133628613 |
- |
- |
PREDICTED: L-Ala-D/L-amino acid epimerase isoform X2 [Jatropha curcas] |
| 14 |
Hb_000816_200 |
0.1164478325 |
- |
- |
2-deoxyglucose-6-phosphate phosphatase, putative [Ricinus communis] |
| 15 |
Hb_000684_430 |
0.1178372052 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g02830, chloroplastic [Jatropha curcas] |
| 16 |
Hb_004871_010 |
0.1179881208 |
- |
- |
RNA polymerase sigma factor rpoD1, putative [Ricinus communis] |
| 17 |
Hb_006829_080 |
0.1180034841 |
- |
- |
PREDICTED: protein LHCP TRANSLOCATION DEFECT [Jatropha curcas] |
| 18 |
Hb_001959_060 |
0.1182133946 |
- |
- |
PREDICTED: carboxyl-terminal-processing peptidase 2, chloroplastic [Jatropha curcas] |
| 19 |
Hb_000085_070 |
0.1192727935 |
- |
- |
PREDICTED: trigger factor-like protein TIG, Chloroplastic isoform X1 [Jatropha curcas] |
| 20 |
Hb_002542_160 |
0.1202802715 |
- |
- |
PREDICTED: peptide chain release factor APG3, chloroplastic [Jatropha curcas] |