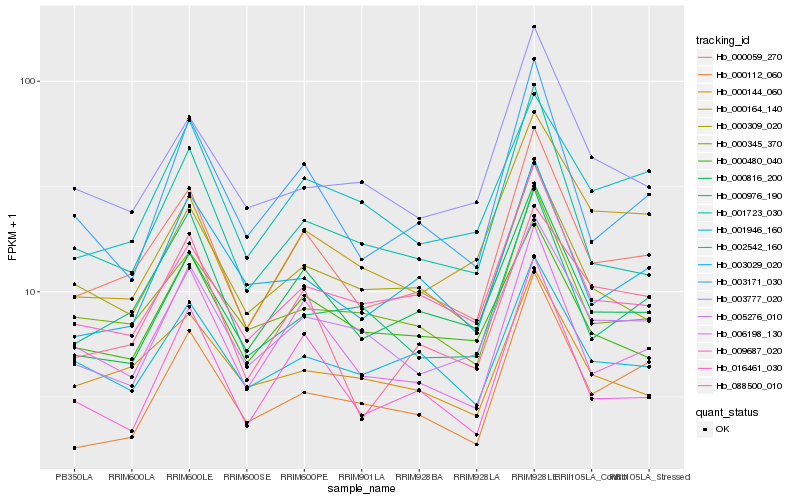
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000059_270 |
0.0 |
- |
- |
superoxide dismutase [Fe], chloroplastic [Jatropha curcas] |
| 2 |
Hb_006198_130 |
0.0824857262 |
- |
- |
PREDICTED: uncharacterized protein LOC105644406 [Jatropha curcas] |
| 3 |
Hb_000816_200 |
0.0903424805 |
- |
- |
2-deoxyglucose-6-phosphate phosphatase, putative [Ricinus communis] |
| 4 |
Hb_016461_030 |
0.0948831235 |
- |
- |
PREDICTED: putative leucine--tRNA ligase, mitochondrial isoform X1 [Jatropha curcas] |
| 5 |
Hb_003171_030 |
0.0949316017 |
- |
- |
PREDICTED: grpE protein homolog, mitochondrial [Jatropha curcas] |
| 6 |
Hb_009687_020 |
0.0967598414 |
- |
- |
hypothetical protein JCGZ_10323 [Jatropha curcas] |
| 7 |
Hb_000345_370 |
0.0995385123 |
- |
- |
PREDICTED: probable Xaa-Pro aminopeptidase P [Jatropha curcas] |
| 8 |
Hb_088500_010 |
0.1017418914 |
- |
- |
PREDICTED: haloacid dehalogenase-like hydrolase domain-containing protein At4g39970 [Jatropha curcas] |
| 9 |
Hb_001723_030 |
0.1025563829 |
- |
- |
PREDICTED: 30S ribosomal protein S1, chloroplastic [Jatropha curcas] |
| 10 |
Hb_005276_010 |
0.1045469461 |
- |
- |
hypothetical protein CICLE_v10021605mg [Citrus clementina] |
| 11 |
Hb_000309_020 |
0.1083899488 |
- |
- |
PREDICTED: uncharacterized protein LOC105641764 [Jatropha curcas] |
| 12 |
Hb_000144_060 |
0.1085814633 |
- |
- |
PREDICTED: uncharacterized protein At5g03900, chloroplastic isoform X1 [Jatropha curcas] |
| 13 |
Hb_002542_160 |
0.1094672676 |
- |
- |
PREDICTED: peptide chain release factor APG3, chloroplastic [Jatropha curcas] |
| 14 |
Hb_001946_160 |
0.1118254821 |
- |
- |
putative chaperon P13.9 [Castanea sativa] |
| 15 |
Hb_000112_060 |
0.1130475967 |
- |
- |
PREDICTED: thioredoxin-like protein CITRX, chloroplastic [Jatropha curcas] |
| 16 |
Hb_000164_140 |
0.1133092241 |
- |
- |
cinnamoyl-CoA reductase, putative [Ricinus communis] |
| 17 |
Hb_000976_190 |
0.1135471725 |
- |
- |
PREDICTED: obg-like ATPase 1 [Jatropha curcas] |
| 18 |
Hb_000480_040 |
0.1137714831 |
- |
- |
PREDICTED: L-Ala-D/L-amino acid epimerase isoform X2 [Jatropha curcas] |
| 19 |
Hb_003777_020 |
0.1140476493 |
- |
- |
Fructose-1,6-bisphosphatase, cytosolic, putative [Ricinus communis] |
| 20 |
Hb_003029_020 |
0.1148675945 |
- |
- |
ATP-dependent Clp protease proteolytic subunit, putative [Ricinus communis] |