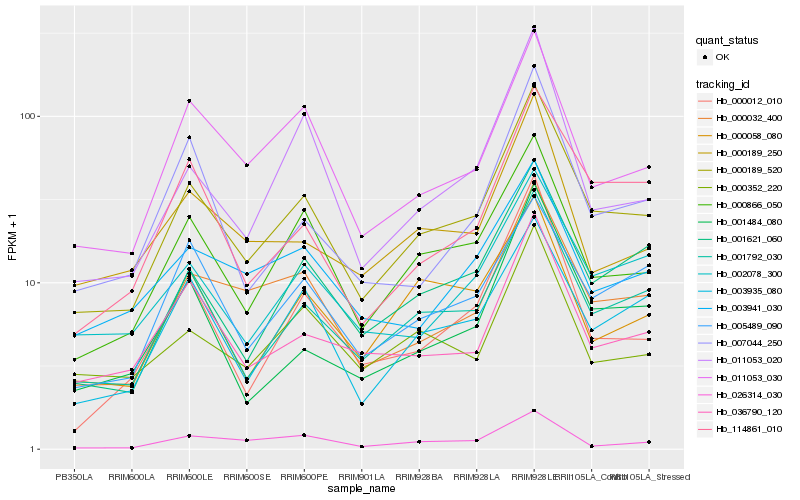
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000189_520 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105647658 [Jatropha curcas] |
| 2 |
Hb_001792_030 |
0.0771550119 |
- |
- |
PREDICTED: aminoacyl tRNA synthase complex-interacting multifunctional protein 1 [Jatropha curcas] |
| 3 |
Hb_000866_050 |
0.0935001879 |
- |
- |
phosphate transporter [Manihot esculenta] |
| 4 |
Hb_000032_400 |
0.1004562375 |
- |
- |
PREDICTED: UDP-glucose 4-epimerase GEPI48 [Jatropha curcas] |
| 5 |
Hb_001621_060 |
0.1055989455 |
- |
- |
PREDICTED: thylakoid lumenal 15 kDa protein 1, chloroplastic [Jatropha curcas] |
| 6 |
Hb_003935_080 |
0.1121973203 |
- |
- |
PREDICTED: mechanosensitive ion channel protein 2, chloroplastic [Jatropha curcas] |
| 7 |
Hb_011053_030 |
0.114229999 |
- |
- |
- |
| 8 |
Hb_002078_300 |
0.1151287108 |
- |
- |
PREDICTED: uncharacterized protein LOC105644817 isoform X1 [Jatropha curcas] |
| 9 |
Hb_114861_010 |
0.1208351819 |
- |
- |
STRESS ENHANCED protein 1 [Populus trichocarpa] |
| 10 |
Hb_036790_120 |
0.121577147 |
- |
- |
PREDICTED: uncharacterized protein LOC105645122 isoform X1 [Jatropha curcas] |
| 11 |
Hb_000012_010 |
0.1272043553 |
- |
- |
hypothetical protein JCGZ_07709 [Jatropha curcas] |
| 12 |
Hb_001484_080 |
0.128014488 |
- |
- |
2-deoxyglucose-6-phosphate phosphatase, putative [Ricinus communis] |
| 13 |
Hb_005489_090 |
0.1302600425 |
- |
- |
PREDICTED: peptidyl-tRNA hydrolase ICT1, mitochondrial [Jatropha curcas] |
| 14 |
Hb_011053_020 |
0.1306555933 |
- |
- |
lipoic acid synthetase, putative [Ricinus communis] |
| 15 |
Hb_000058_080 |
0.1320901583 |
- |
- |
PREDICTED: phenylalanine--tRNA ligase, chloroplastic/mitochondrial [Jatropha curcas] |
| 16 |
Hb_000352_220 |
0.1325180454 |
- |
- |
PREDICTED: probable tyrosine--tRNA ligase, mitochondrial [Jatropha curcas] |
| 17 |
Hb_000189_250 |
0.1327542277 |
- |
- |
PREDICTED: probable ribose-5-phosphate isomerase 3, chloroplastic [Jatropha curcas] |
| 18 |
Hb_003941_030 |
0.1339973468 |
- |
- |
Protease ecfE, putative [Ricinus communis] |
| 19 |
Hb_026314_030 |
0.1351929041 |
- |
- |
PREDICTED: transmembrane protein 41B [Jatropha curcas] |
| 20 |
Hb_007044_250 |
0.1364460603 |
- |
- |
conserved hypothetical protein [Ricinus communis] |