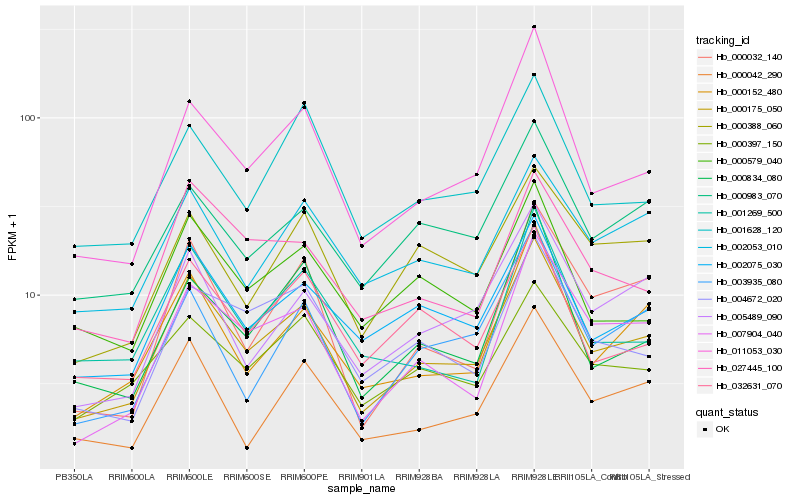
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000175_050 |
0.0 |
- |
- |
- |
| 2 |
Hb_011053_030 |
0.0887926246 |
- |
- |
- |
| 3 |
Hb_007904_040 |
0.0961021108 |
- |
- |
PREDICTED: uncharacterized protein LOC105644727 [Jatropha curcas] |
| 4 |
Hb_000152_480 |
0.0972299787 |
- |
- |
PREDICTED: protein LOW PSII ACCUMULATION 2, chloroplastic [Jatropha curcas] |
| 5 |
Hb_002075_030 |
0.1007361112 |
- |
- |
PREDICTED: glycerol-3-phosphate acyltransferase, chloroplastic [Jatropha curcas] |
| 6 |
Hb_005489_090 |
0.1086761452 |
- |
- |
PREDICTED: peptidyl-tRNA hydrolase ICT1, mitochondrial [Jatropha curcas] |
| 7 |
Hb_000032_140 |
0.1129148715 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_004672_020 |
0.1129549949 |
- |
- |
PREDICTED: ATP-dependent zinc metalloprotease FtsH [Jatropha curcas] |
| 9 |
Hb_000397_150 |
0.1129787204 |
- |
- |
PREDICTED: probable flavin-containing monooxygenase 1 [Jatropha curcas] |
| 10 |
Hb_032631_070 |
0.1155806532 |
- |
- |
PREDICTED: lisH domain and HEAT repeat-containing protein KIAA1468 homolog isoform X1 [Jatropha curcas] |
| 11 |
Hb_000834_080 |
0.1159172366 |
- |
- |
PREDICTED: uncharacterized protein LOC105637718 [Jatropha curcas] |
| 12 |
Hb_000042_290 |
0.1170314735 |
- |
- |
PREDICTED: uncharacterized protein LOC105632818 isoform X2 [Jatropha curcas] |
| 13 |
Hb_000983_070 |
0.1178048227 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_003935_080 |
0.1189474001 |
- |
- |
PREDICTED: mechanosensitive ion channel protein 2, chloroplastic [Jatropha curcas] |
| 15 |
Hb_000579_040 |
0.1199616453 |
- |
- |
PREDICTED: putative GTP-binding protein 6 [Jatropha curcas] |
| 16 |
Hb_001269_500 |
0.120377122 |
- |
- |
PREDICTED: protein LOW PSII ACCUMULATION 3, chloroplastic-like isoform X2 [Populus euphratica] |
| 17 |
Hb_001628_120 |
0.1215389439 |
- |
- |
putative ascorbate peroxidase, partial [Taraxacum brevicorniculatum] |
| 18 |
Hb_002053_010 |
0.1229086183 |
- |
- |
PREDICTED: clp protease-related protein At4g12060, chloroplastic-like [Jatropha curcas] |
| 19 |
Hb_000388_060 |
0.1232525829 |
- |
- |
fructokinase [Manihot esculenta] |
| 20 |
Hb_027445_100 |
0.1233365918 |
- |
- |
PREDICTED: probable monodehydroascorbate reductase, cytoplasmic isoform 2 [Jatropha curcas] |