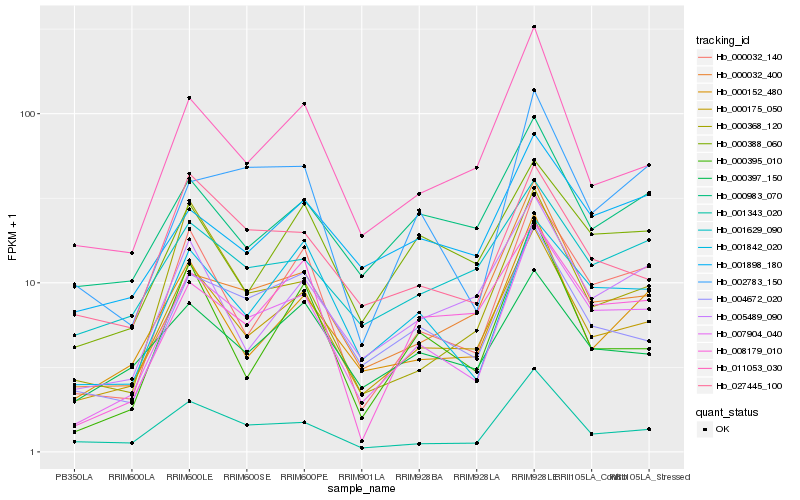
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007904_040 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105644727 [Jatropha curcas] |
| 2 |
Hb_000175_050 |
0.0961021108 |
- |
- |
- |
| 3 |
Hb_000032_140 |
0.1075370721 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_001842_020 |
0.1206319536 |
- |
- |
PREDICTED: low molecular weight phosphotyrosine protein phosphatase [Jatropha curcas] |
| 5 |
Hb_004672_020 |
0.1248861332 |
- |
- |
PREDICTED: ATP-dependent zinc metalloprotease FtsH [Jatropha curcas] |
| 6 |
Hb_000032_400 |
0.1276103972 |
- |
- |
PREDICTED: UDP-glucose 4-epimerase GEPI48 [Jatropha curcas] |
| 7 |
Hb_002783_150 |
0.1357167397 |
- |
- |
cyclophilin [Hevea brasiliensis] |
| 8 |
Hb_001343_020 |
0.1365042102 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 9 |
Hb_000152_480 |
0.1407056388 |
- |
- |
PREDICTED: protein LOW PSII ACCUMULATION 2, chloroplastic [Jatropha curcas] |
| 10 |
Hb_011053_030 |
0.1419066903 |
- |
- |
- |
| 11 |
Hb_000388_060 |
0.148163073 |
- |
- |
fructokinase [Manihot esculenta] |
| 12 |
Hb_008179_010 |
0.1503968886 |
transcription factor |
TF Family: ARF |
PREDICTED: auxin response factor 16 [Jatropha curcas] |
| 13 |
Hb_005489_090 |
0.1509352176 |
- |
- |
PREDICTED: peptidyl-tRNA hydrolase ICT1, mitochondrial [Jatropha curcas] |
| 14 |
Hb_001898_180 |
0.1535274938 |
- |
- |
PREDICTED: translation initiation factor IF-1, chloroplastic [Jatropha curcas] |
| 15 |
Hb_027445_100 |
0.1535526768 |
- |
- |
PREDICTED: probable monodehydroascorbate reductase, cytoplasmic isoform 2 [Jatropha curcas] |
| 16 |
Hb_000395_010 |
0.1543932424 |
- |
- |
alpha/beta hydrolase domain containing protein 1,3, putative [Ricinus communis] |
| 17 |
Hb_001629_090 |
0.1549188593 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_000368_120 |
0.1554284656 |
- |
- |
PREDICTED: uncharacterized protein LOC105633862 [Jatropha curcas] |
| 19 |
Hb_000397_150 |
0.1556891787 |
- |
- |
PREDICTED: probable flavin-containing monooxygenase 1 [Jatropha curcas] |
| 20 |
Hb_000983_070 |
0.1564263206 |
- |
- |
conserved hypothetical protein [Ricinus communis] |