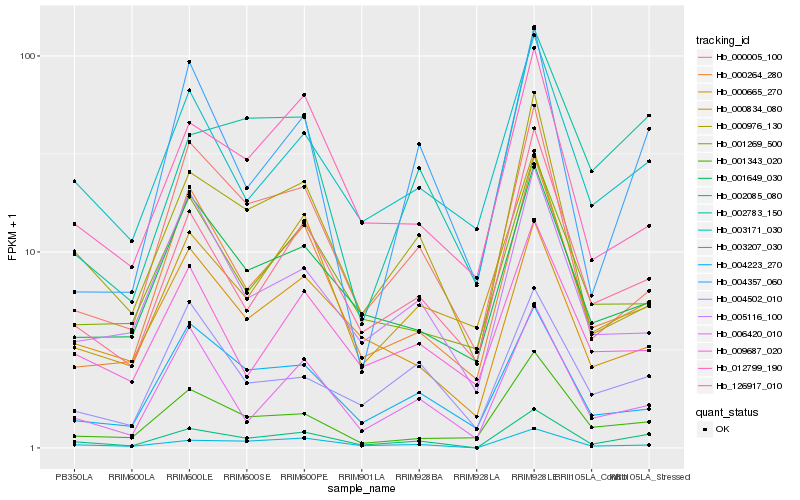
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002085_080 |
0.0 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 68-like [Jatropha curcas] |
| 2 |
Hb_003207_030 |
0.1341334349 |
- |
- |
hypothetical protein VITISV_027754 [Vitis vinifera] |
| 3 |
Hb_126917_010 |
0.1472180232 |
- |
- |
PREDICTED: uncharacterized protein LOC105646333 [Jatropha curcas] |
| 4 |
Hb_003171_030 |
0.1635294819 |
- |
- |
PREDICTED: grpE protein homolog, mitochondrial [Jatropha curcas] |
| 5 |
Hb_002783_150 |
0.1676610339 |
- |
- |
cyclophilin [Hevea brasiliensis] |
| 6 |
Hb_000264_280 |
0.1695211802 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RMA1H1-like [Jatropha curcas] |
| 7 |
Hb_000976_130 |
0.1700781101 |
- |
- |
PREDICTED: probable BOI-related E3 ubiquitin-protein ligase 2 [Jatropha curcas] |
| 8 |
Hb_006420_010 |
0.1711543207 |
- |
- |
PREDICTED: uncharacterized protein LOC105649815 [Jatropha curcas] |
| 9 |
Hb_000665_270 |
0.1729914581 |
- |
- |
PREDICTED: adenine phosphoribosyltransferase 5 [Jatropha curcas] |
| 10 |
Hb_001343_020 |
0.174161789 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 11 |
Hb_001649_030 |
0.1750148471 |
- |
- |
PREDICTED: peptide deformylase 1B, chloroplastic [Jatropha curcas] |
| 12 |
Hb_000005_100 |
0.1767048323 |
- |
- |
PREDICTED: pheophorbide a oxygenase, chloroplastic [Jatropha curcas] |
| 13 |
Hb_004357_060 |
0.1769131942 |
- |
- |
PREDICTED: uncharacterized protein At4g22758 [Jatropha curcas] |
| 14 |
Hb_000834_080 |
0.17873497 |
- |
- |
PREDICTED: uncharacterized protein LOC105637718 [Jatropha curcas] |
| 15 |
Hb_012799_190 |
0.179594981 |
- |
- |
PREDICTED: uncharacterized protein LOC105648490 [Jatropha curcas] |
| 16 |
Hb_004223_270 |
0.1815164621 |
- |
- |
PREDICTED: uncharacterized protein LOC105635371 isoform X1 [Jatropha curcas] |
| 17 |
Hb_001269_500 |
0.1829404546 |
- |
- |
PREDICTED: protein LOW PSII ACCUMULATION 3, chloroplastic-like isoform X2 [Populus euphratica] |
| 18 |
Hb_004502_010 |
0.1837267876 |
- |
- |
hypothetical protein JCGZ_23576 [Jatropha curcas] |
| 19 |
Hb_009687_020 |
0.1855079206 |
- |
- |
hypothetical protein JCGZ_10323 [Jatropha curcas] |
| 20 |
Hb_005116_100 |
0.1867337692 |
- |
- |
PREDICTED: uncharacterized protein LOC105649999 [Jatropha curcas] |