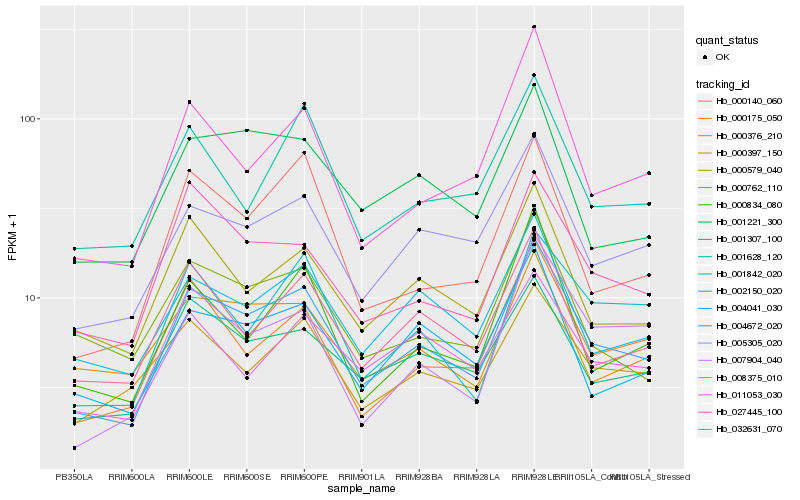
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004672_020 |
0.0 |
- |
- |
PREDICTED: ATP-dependent zinc metalloprotease FtsH [Jatropha curcas] |
| 2 |
Hb_005305_020 |
0.1009136911 |
- |
- |
PREDICTED: dicarboxylate transporter 1, chloroplastic [Jatropha curcas] |
| 3 |
Hb_001307_100 |
0.1059948519 |
- |
- |
calmodulin binding protein, putative [Ricinus communis] |
| 4 |
Hb_004041_030 |
0.1100934783 |
- |
- |
PREDICTED: mitochondrial substrate carrier family protein C-like isoform X1 [Populus euphratica] |
| 5 |
Hb_002150_020 |
0.1111194207 |
- |
- |
PREDICTED: translation initiation factor IF-2, mitochondrial isoform X1 [Jatropha curcas] |
| 6 |
Hb_032631_070 |
0.1123571583 |
- |
- |
PREDICTED: lisH domain and HEAT repeat-containing protein KIAA1468 homolog isoform X1 [Jatropha curcas] |
| 7 |
Hb_000175_050 |
0.1129549949 |
- |
- |
- |
| 8 |
Hb_000579_040 |
0.1139230792 |
- |
- |
PREDICTED: putative GTP-binding protein 6 [Jatropha curcas] |
| 9 |
Hb_000397_150 |
0.1180069535 |
- |
- |
PREDICTED: probable flavin-containing monooxygenase 1 [Jatropha curcas] |
| 10 |
Hb_027445_100 |
0.1198773153 |
- |
- |
PREDICTED: probable monodehydroascorbate reductase, cytoplasmic isoform 2 [Jatropha curcas] |
| 11 |
Hb_000140_060 |
0.1205148535 |
- |
- |
50S ribosomal protein L5, putative [Ricinus communis] |
| 12 |
Hb_001221_300 |
0.1223822281 |
- |
- |
PREDICTED: acetolactate synthase 3, chloroplastic [Jatropha curcas] |
| 13 |
Hb_000834_080 |
0.1227900247 |
- |
- |
PREDICTED: uncharacterized protein LOC105637718 [Jatropha curcas] |
| 14 |
Hb_001842_020 |
0.1241399379 |
- |
- |
PREDICTED: low molecular weight phosphotyrosine protein phosphatase [Jatropha curcas] |
| 15 |
Hb_011053_030 |
0.1241830404 |
- |
- |
- |
| 16 |
Hb_007904_040 |
0.1248861332 |
- |
- |
PREDICTED: uncharacterized protein LOC105644727 [Jatropha curcas] |
| 17 |
Hb_001628_120 |
0.1264047957 |
- |
- |
putative ascorbate peroxidase, partial [Taraxacum brevicorniculatum] |
| 18 |
Hb_000762_110 |
0.1266867026 |
- |
- |
PREDICTED: protease Do-like 1, chloroplastic [Jatropha curcas] |
| 19 |
Hb_000376_210 |
0.1270434219 |
- |
- |
PREDICTED: O-acyltransferase WSD1-like [Jatropha curcas] |
| 20 |
Hb_008375_010 |
0.1302260057 |
- |
- |
PREDICTED: carbon catabolite repressor protein 4 homolog 4 isoform X3 [Pyrus x bretschneideri] |