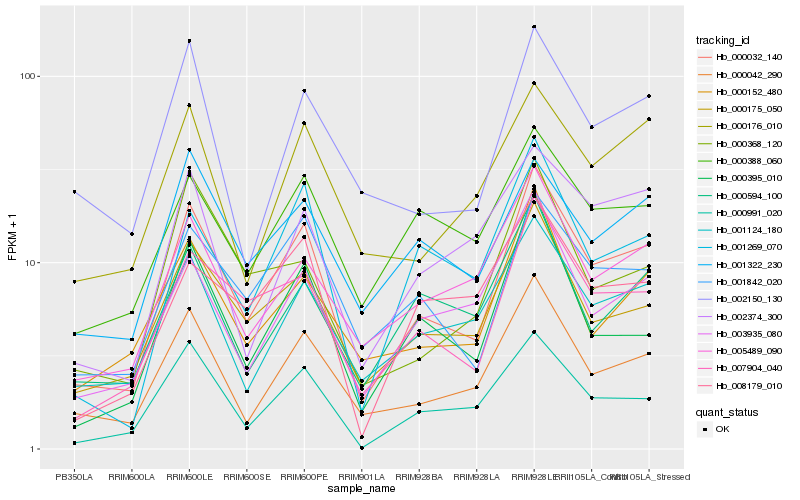
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000032_140 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_007904_040 |
0.1075370721 |
- |
- |
PREDICTED: uncharacterized protein LOC105644727 [Jatropha curcas] |
| 3 |
Hb_000042_290 |
0.1100380744 |
- |
- |
PREDICTED: uncharacterized protein LOC105632818 isoform X2 [Jatropha curcas] |
| 4 |
Hb_000175_050 |
0.1129148715 |
- |
- |
- |
| 5 |
Hb_000991_020 |
0.1199878425 |
- |
- |
amino acid transporter, putative [Ricinus communis] |
| 6 |
Hb_001842_020 |
0.1241490904 |
- |
- |
PREDICTED: low molecular weight phosphotyrosine protein phosphatase [Jatropha curcas] |
| 7 |
Hb_001124_180 |
0.1292869658 |
- |
- |
PREDICTED: uncharacterized protein LOC105171432 [Sesamum indicum] |
| 8 |
Hb_003935_080 |
0.131668195 |
- |
- |
PREDICTED: mechanosensitive ion channel protein 2, chloroplastic [Jatropha curcas] |
| 9 |
Hb_005489_090 |
0.1343882422 |
- |
- |
PREDICTED: peptidyl-tRNA hydrolase ICT1, mitochondrial [Jatropha curcas] |
| 10 |
Hb_000395_010 |
0.1347390494 |
- |
- |
alpha/beta hydrolase domain containing protein 1,3, putative [Ricinus communis] |
| 11 |
Hb_000152_480 |
0.1357733494 |
- |
- |
PREDICTED: protein LOW PSII ACCUMULATION 2, chloroplastic [Jatropha curcas] |
| 12 |
Hb_001269_070 |
0.1363652417 |
- |
- |
double-stranded RNA binding protein, putative [Ricinus communis] |
| 13 |
Hb_000176_010 |
0.1442768135 |
- |
- |
PREDICTED: 50S ribosomal protein L35, chloroplastic [Jatropha curcas] |
| 14 |
Hb_002150_130 |
0.1466869028 |
- |
- |
50S ribosomal protein L18, chloroplast precursor, putative [Ricinus communis] |
| 15 |
Hb_001322_230 |
0.1472046788 |
- |
- |
PREDICTED: ATP-dependent Clp protease proteolytic subunit-related protein 4, chloroplastic [Populus euphratica] |
| 16 |
Hb_000388_060 |
0.1482558388 |
- |
- |
fructokinase [Manihot esculenta] |
| 17 |
Hb_000594_100 |
0.1491047004 |
- |
- |
PREDICTED: methionine aminopeptidase 1B, chloroplastic [Jatropha curcas] |
| 18 |
Hb_002374_300 |
0.1495481634 |
- |
- |
hypothetical protein JCGZ_12140 [Jatropha curcas] |
| 19 |
Hb_008179_010 |
0.1520967753 |
transcription factor |
TF Family: ARF |
PREDICTED: auxin response factor 16 [Jatropha curcas] |
| 20 |
Hb_000368_120 |
0.1532014698 |
- |
- |
PREDICTED: uncharacterized protein LOC105633862 [Jatropha curcas] |