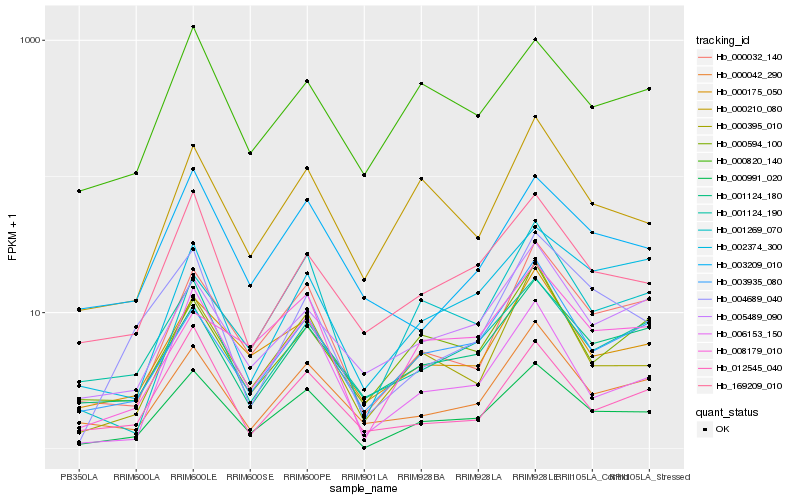
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000991_020 |
0.0 |
- |
- |
amino acid transporter, putative [Ricinus communis] |
| 2 |
Hb_000032_140 |
0.1199878425 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_000395_010 |
0.1312167933 |
- |
- |
alpha/beta hydrolase domain containing protein 1,3, putative [Ricinus communis] |
| 4 |
Hb_000175_050 |
0.135732468 |
- |
- |
- |
| 5 |
Hb_000042_290 |
0.1383165972 |
- |
- |
PREDICTED: uncharacterized protein LOC105632818 isoform X2 [Jatropha curcas] |
| 6 |
Hb_169209_010 |
0.1395757606 |
- |
- |
PREDICTED: porphobilinogen deaminase, chloroplastic [Jatropha curcas] |
| 7 |
Hb_001124_180 |
0.1423496804 |
- |
- |
PREDICTED: uncharacterized protein LOC105171432 [Sesamum indicum] |
| 8 |
Hb_003935_080 |
0.1456685991 |
- |
- |
PREDICTED: mechanosensitive ion channel protein 2, chloroplastic [Jatropha curcas] |
| 9 |
Hb_000210_080 |
0.14900795 |
- |
- |
PREDICTED: 2-Cys peroxiredoxin BAS1, chloroplastic-like [Jatropha curcas] |
| 10 |
Hb_001124_190 |
0.1533038928 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_000820_140 |
0.1542768384 |
- |
- |
histone H4 [Zea mays] |
| 12 |
Hb_008179_010 |
0.1547136818 |
transcription factor |
TF Family: ARF |
PREDICTED: auxin response factor 16 [Jatropha curcas] |
| 13 |
Hb_002374_300 |
0.1559869856 |
- |
- |
hypothetical protein JCGZ_12140 [Jatropha curcas] |
| 14 |
Hb_006153_150 |
0.1560900566 |
- |
- |
PREDICTED: uncharacterized protein LOC105645999 [Jatropha curcas] |
| 15 |
Hb_005489_090 |
0.1575114771 |
- |
- |
PREDICTED: peptidyl-tRNA hydrolase ICT1, mitochondrial [Jatropha curcas] |
| 16 |
Hb_004689_040 |
0.1579592882 |
- |
- |
PREDICTED: uncharacterized protein LOC105648084 [Jatropha curcas] |
| 17 |
Hb_001269_070 |
0.1605013824 |
- |
- |
double-stranded RNA binding protein, putative [Ricinus communis] |
| 18 |
Hb_003209_010 |
0.1614255319 |
- |
- |
hypothetical protein POPTR_0010s16880g [Populus trichocarpa] |
| 19 |
Hb_000594_100 |
0.1631714467 |
- |
- |
PREDICTED: methionine aminopeptidase 1B, chloroplastic [Jatropha curcas] |
| 20 |
Hb_012545_040 |
0.1640973561 |
- |
- |
PREDICTED: uroporphyrinogen-III synthase, chloroplastic isoform X1 [Jatropha curcas] |