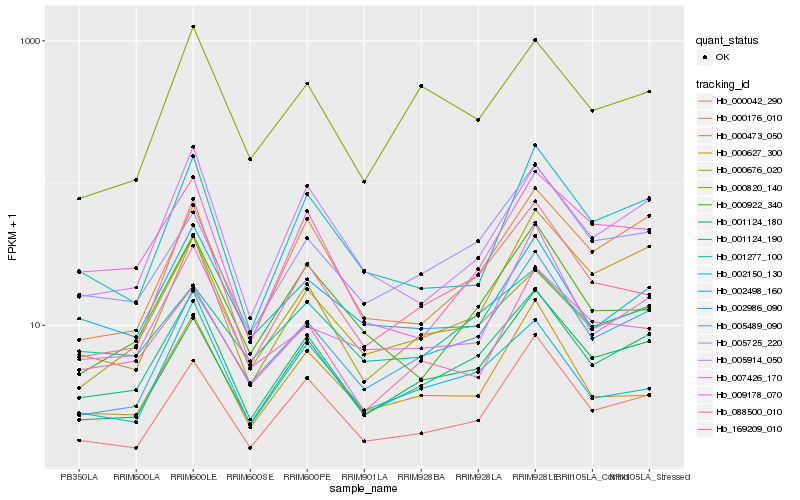
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001124_190 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_001124_180 |
0.0952604983 |
- |
- |
PREDICTED: uncharacterized protein LOC105171432 [Sesamum indicum] |
| 3 |
Hb_169209_010 |
0.1079153422 |
- |
- |
PREDICTED: porphobilinogen deaminase, chloroplastic [Jatropha curcas] |
| 4 |
Hb_005914_050 |
0.1108391578 |
- |
- |
PREDICTED: 50S ribosomal protein L28, chloroplastic [Jatropha curcas] |
| 5 |
Hb_000042_290 |
0.1124099654 |
- |
- |
PREDICTED: uncharacterized protein LOC105632818 isoform X2 [Jatropha curcas] |
| 6 |
Hb_000676_020 |
0.1129989907 |
- |
- |
PREDICTED: uncharacterized protein LOC105645386 [Jatropha curcas] |
| 7 |
Hb_000176_010 |
0.1163794851 |
- |
- |
PREDICTED: 50S ribosomal protein L35, chloroplastic [Jatropha curcas] |
| 8 |
Hb_009178_070 |
0.1180513369 |
- |
- |
PREDICTED: probable plastid-lipid-associated protein 8, chloroplastic [Jatropha curcas] |
| 9 |
Hb_002986_090 |
0.12163622 |
- |
- |
PREDICTED: methionine aminopeptidase 1D, chloroplastic/mitochondrial [Jatropha curcas] |
| 10 |
Hb_002150_130 |
0.1218646511 |
- |
- |
50S ribosomal protein L18, chloroplast precursor, putative [Ricinus communis] |
| 11 |
Hb_005489_090 |
0.12372814 |
- |
- |
PREDICTED: peptidyl-tRNA hydrolase ICT1, mitochondrial [Jatropha curcas] |
| 12 |
Hb_088500_010 |
0.1238955251 |
- |
- |
PREDICTED: haloacid dehalogenase-like hydrolase domain-containing protein At4g39970 [Jatropha curcas] |
| 13 |
Hb_000922_340 |
0.1248111407 |
- |
- |
PREDICTED: uncharacterized protein LOC105640368 [Jatropha curcas] |
| 14 |
Hb_002498_160 |
0.1260239653 |
- |
- |
PREDICTED: uncharacterized protein LOC105649315 [Jatropha curcas] |
| 15 |
Hb_005725_220 |
0.127390057 |
- |
- |
PREDICTED: uncharacterized protein LOC105630808 [Jatropha curcas] |
| 16 |
Hb_001277_100 |
0.1295925343 |
- |
- |
PREDICTED: 2-methyl-6-phytyl-1,4-hydroquinone methyltransferase, chloroplastic-like isoform X1 [Gossypium raimondii] |
| 17 |
Hb_000473_050 |
0.1298584642 |
- |
- |
chromatin regulatory protein sir2, putative [Ricinus communis] |
| 18 |
Hb_007426_170 |
0.1309601682 |
transcription factor |
TF Family: TCP |
- |
| 19 |
Hb_000820_140 |
0.1310513308 |
- |
- |
histone H4 [Zea mays] |
| 20 |
Hb_000627_300 |
0.1319286114 |
- |
- |
PREDICTED: uncharacterized protein LOC105632672 [Jatropha curcas] |