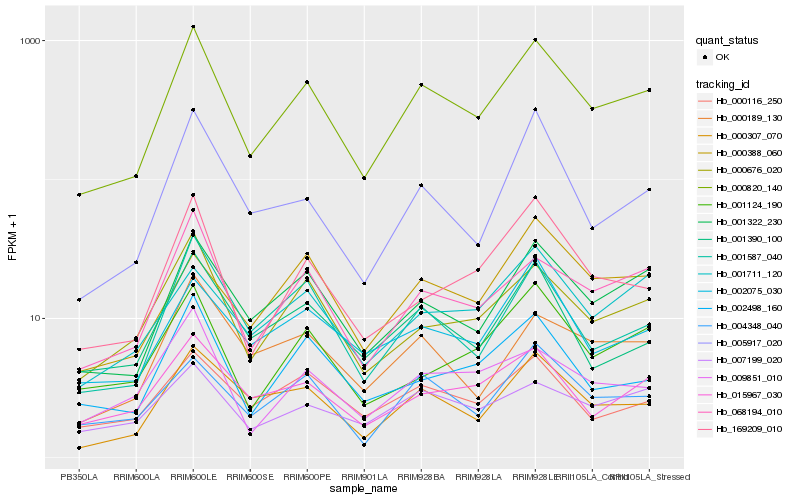
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000820_140 |
0.0 |
- |
- |
histone H4 [Zea mays] |
| 2 |
Hb_001322_230 |
0.0921645642 |
- |
- |
PREDICTED: ATP-dependent Clp protease proteolytic subunit-related protein 4, chloroplastic [Populus euphratica] |
| 3 |
Hb_068194_010 |
0.1004735782 |
- |
- |
hypothetical protein POPTR_0010s23740g [Populus trichocarpa] |
| 4 |
Hb_000307_070 |
0.1099473494 |
- |
- |
protein kinase, putative [Ricinus communis] |
| 5 |
Hb_000676_020 |
0.1101443128 |
- |
- |
PREDICTED: uncharacterized protein LOC105645386 [Jatropha curcas] |
| 6 |
Hb_001587_040 |
0.1162594305 |
- |
- |
PREDICTED: soluble inorganic pyrophosphatase 6, chloroplastic [Jatropha curcas] |
| 7 |
Hb_004348_040 |
0.122774963 |
- |
- |
type I inositol polyphosphate 5-phosphatase, putative [Ricinus communis] |
| 8 |
Hb_015967_030 |
0.1280589137 |
transcription factor |
TF Family: B3 |
PREDICTED: B3 domain-containing transcription repressor VAL1 isoform X1 [Jatropha curcas] |
| 9 |
Hb_000116_250 |
0.1288092266 |
- |
- |
PREDICTED: DNA mismatch repair protein MLH3 [Jatropha curcas] |
| 10 |
Hb_169209_010 |
0.1290334333 |
- |
- |
PREDICTED: porphobilinogen deaminase, chloroplastic [Jatropha curcas] |
| 11 |
Hb_001390_100 |
0.129049487 |
- |
- |
PREDICTED: LL-diaminopimelate aminotransferase, chloroplastic [Jatropha curcas] |
| 12 |
Hb_002498_160 |
0.1293288287 |
- |
- |
PREDICTED: uncharacterized protein LOC105649315 [Jatropha curcas] |
| 13 |
Hb_000189_130 |
0.1305563921 |
- |
- |
PREDICTED: sec-independent protein translocase protein TATA, chloroplastic [Jatropha curcas] |
| 14 |
Hb_001124_190 |
0.1310513308 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_002075_030 |
0.1324738333 |
- |
- |
PREDICTED: glycerol-3-phosphate acyltransferase, chloroplastic [Jatropha curcas] |
| 16 |
Hb_007199_020 |
0.1331379489 |
- |
- |
hypothetical protein CICLE_v10028570mg [Citrus clementina] |
| 17 |
Hb_005917_020 |
0.1348412318 |
- |
- |
Winged-helix DNA-binding transcription factor family protein [Theobroma cacao] |
| 18 |
Hb_009851_010 |
0.1364994218 |
- |
- |
PREDICTED: beta-amylase 3, chloroplastic-like [Jatropha curcas] |
| 19 |
Hb_001711_120 |
0.1367165007 |
- |
- |
PREDICTED: ATP-dependent Clp protease proteolytic subunit 4, chloroplastic [Jatropha curcas] |
| 20 |
Hb_000388_060 |
0.1396893771 |
- |
- |
fructokinase [Manihot esculenta] |