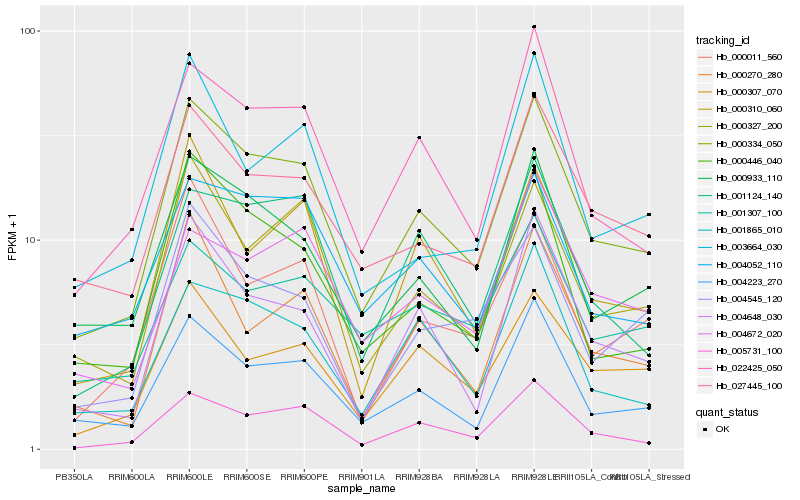
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000334_050 |
0.0 |
- |
- |
PREDICTED: vesicle-associated protein 4-1-like [Jatropha curcas] |
| 2 |
Hb_004223_270 |
0.0871898019 |
- |
- |
PREDICTED: uncharacterized protein LOC105635371 isoform X1 [Jatropha curcas] |
| 3 |
Hb_022425_050 |
0.09317242 |
- |
- |
PREDICTED: probable lactoylglutathione lyase, chloroplast isoform X1 [Jatropha curcas] |
| 4 |
Hb_000933_110 |
0.0938151121 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 3 [Jatropha curcas] |
| 5 |
Hb_027445_100 |
0.098015276 |
- |
- |
PREDICTED: probable monodehydroascorbate reductase, cytoplasmic isoform 2 [Jatropha curcas] |
| 6 |
Hb_005731_100 |
0.1007230826 |
- |
- |
monovalent cation:proton antiporter, putative [Ricinus communis] |
| 7 |
Hb_000327_200 |
0.105709929 |
- |
- |
glutathione-s-transferase omega, putative [Ricinus communis] |
| 8 |
Hb_000446_040 |
0.1071786737 |
- |
- |
PREDICTED: uncharacterized protein ycf45 [Jatropha curcas] |
| 9 |
Hb_000307_070 |
0.1111700636 |
- |
- |
protein kinase, putative [Ricinus communis] |
| 10 |
Hb_004648_030 |
0.115342198 |
- |
- |
PREDICTED: probable dimethyladenosine transferase [Jatropha curcas] |
| 11 |
Hb_004052_110 |
0.1155650889 |
- |
- |
PREDICTED: host cell factor 2 isoform X1 [Jatropha curcas] |
| 12 |
Hb_001865_010 |
0.1164334887 |
transcription factor |
TF Family: B3 |
hypothetical protein RCOM_0465210 [Ricinus communis] |
| 13 |
Hb_001124_140 |
0.1185033536 |
- |
- |
PREDICTED: inactive beta-amylase 9 [Jatropha curcas] |
| 14 |
Hb_000310_060 |
0.1202459685 |
- |
- |
hypothetical protein JCGZ_20793 [Jatropha curcas] |
| 15 |
Hb_004545_120 |
0.1210779757 |
- |
- |
PREDICTED: uncharacterized protein LOC105647604 [Jatropha curcas] |
| 16 |
Hb_000270_280 |
0.1254836348 |
- |
- |
Rhicadhesin receptor precursor, putative [Ricinus communis] |
| 17 |
Hb_000011_560 |
0.1256556726 |
- |
- |
PREDICTED: phosphoinositide phosphatase SAC2 isoform X1 [Jatropha curcas] |
| 18 |
Hb_003664_030 |
0.1281321555 |
- |
- |
PREDICTED: mitochondrial carnitine/acylcarnitine carrier-like protein [Jatropha curcas] |
| 19 |
Hb_001307_100 |
0.1291435509 |
- |
- |
calmodulin binding protein, putative [Ricinus communis] |
| 20 |
Hb_004672_020 |
0.1303895342 |
- |
- |
PREDICTED: ATP-dependent zinc metalloprotease FtsH [Jatropha curcas] |