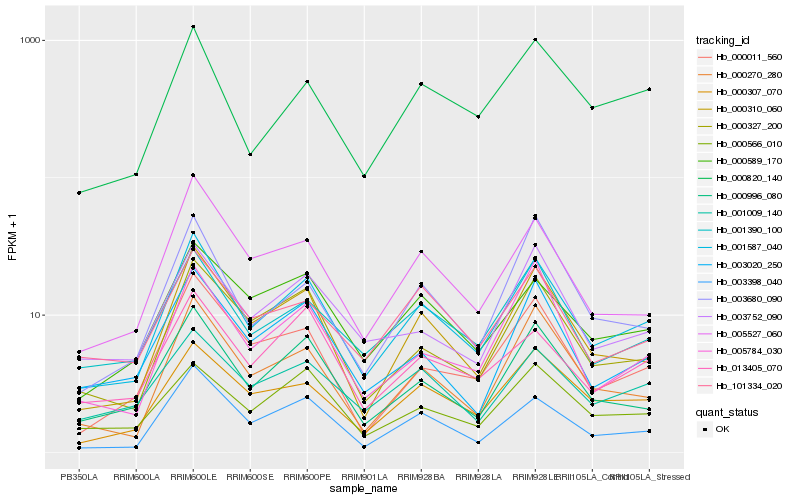
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001587_040 |
0.0 |
- |
- |
PREDICTED: soluble inorganic pyrophosphatase 6, chloroplastic [Jatropha curcas] |
| 2 |
Hb_000310_060 |
0.0879256111 |
- |
- |
hypothetical protein JCGZ_20793 [Jatropha curcas] |
| 3 |
Hb_003398_040 |
0.0959425192 |
- |
- |
PREDICTED: actin-related protein 5 isoform X1 [Jatropha curcas] |
| 4 |
Hb_005527_060 |
0.0998127227 |
- |
- |
malic enzyme, putative [Ricinus communis] |
| 5 |
Hb_000996_080 |
0.0999706693 |
- |
- |
PREDICTED: intracellular protein transport protein USO1 [Jatropha curcas] |
| 6 |
Hb_000307_070 |
0.1011936729 |
- |
- |
protein kinase, putative [Ricinus communis] |
| 7 |
Hb_001390_100 |
0.1014702004 |
- |
- |
PREDICTED: LL-diaminopimelate aminotransferase, chloroplastic [Jatropha curcas] |
| 8 |
Hb_000327_200 |
0.1029559619 |
- |
- |
glutathione-s-transferase omega, putative [Ricinus communis] |
| 9 |
Hb_000270_280 |
0.1093081967 |
- |
- |
Rhicadhesin receptor precursor, putative [Ricinus communis] |
| 10 |
Hb_000589_170 |
0.1120228804 |
- |
- |
PREDICTED: telomere repeat-binding protein 4 isoform X3 [Jatropha curcas] |
| 11 |
Hb_001009_140 |
0.1127217858 |
- |
- |
hypothetical protein JCGZ_11337 [Jatropha curcas] |
| 12 |
Hb_000011_560 |
0.112911865 |
- |
- |
PREDICTED: phosphoinositide phosphatase SAC2 isoform X1 [Jatropha curcas] |
| 13 |
Hb_003020_250 |
0.1141920701 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase PP1 isozyme 2 [Jatropha curcas] |
| 14 |
Hb_013405_070 |
0.115507874 |
- |
- |
ferric-chelate reductase, putative [Ricinus communis] |
| 15 |
Hb_000820_140 |
0.1162594305 |
- |
- |
histone H4 [Zea mays] |
| 16 |
Hb_000566_010 |
0.1199544424 |
- |
- |
glucose-6-phosphate dehydrogenase [Hevea brasiliensis] |
| 17 |
Hb_003752_090 |
0.1210123852 |
- |
- |
chitinase, putative [Ricinus communis] |
| 18 |
Hb_101334_020 |
0.1225769839 |
- |
- |
PREDICTED: uncharacterized WD repeat-containing protein C2A9.03-like [Jatropha curcas] |
| 19 |
Hb_005784_030 |
0.1232025333 |
- |
- |
PREDICTED: uncharacterized protein LOC105645162 isoform X2 [Jatropha curcas] |
| 20 |
Hb_003680_090 |
0.1235734735 |
- |
- |
PREDICTED: threonine dehydratase biosynthetic, chloroplastic [Jatropha curcas] |