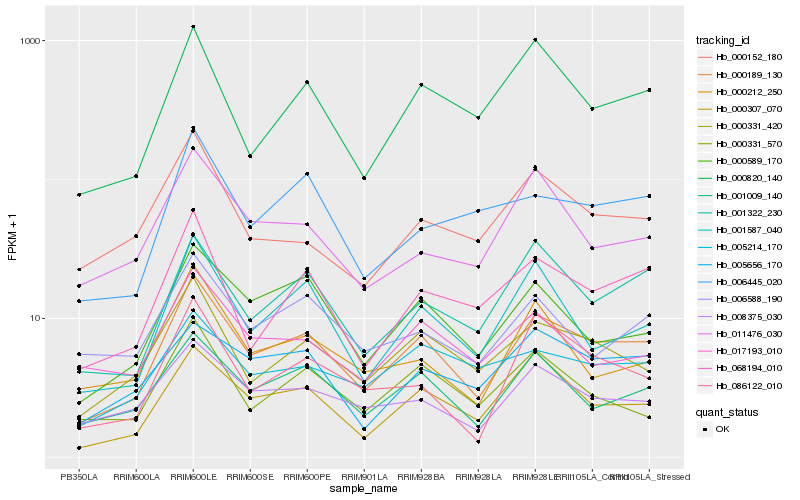
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000189_130 |
0.0 |
- |
- |
PREDICTED: sec-independent protein translocase protein TATA, chloroplastic [Jatropha curcas] |
| 2 |
Hb_000307_070 |
0.1129255846 |
- |
- |
protein kinase, putative [Ricinus communis] |
| 3 |
Hb_001009_140 |
0.1160111062 |
- |
- |
hypothetical protein JCGZ_11337 [Jatropha curcas] |
| 4 |
Hb_000331_420 |
0.1160462769 |
- |
- |
PREDICTED: nucleoside diphosphate kinase 2, chloroplastic [Jatropha curcas] |
| 5 |
Hb_000589_170 |
0.124003284 |
- |
- |
PREDICTED: telomere repeat-binding protein 4 isoform X3 [Jatropha curcas] |
| 6 |
Hb_001587_040 |
0.1247419653 |
- |
- |
PREDICTED: soluble inorganic pyrophosphatase 6, chloroplastic [Jatropha curcas] |
| 7 |
Hb_001322_230 |
0.1261368781 |
- |
- |
PREDICTED: ATP-dependent Clp protease proteolytic subunit-related protein 4, chloroplastic [Populus euphratica] |
| 8 |
Hb_008375_030 |
0.1271239084 |
- |
- |
PREDICTED: carbon catabolite repressor protein 4 homolog 4 isoform X1 [Populus euphratica] |
| 9 |
Hb_068194_010 |
0.1273354901 |
- |
- |
hypothetical protein POPTR_0010s23740g [Populus trichocarpa] |
| 10 |
Hb_000820_140 |
0.1305563921 |
- |
- |
histone H4 [Zea mays] |
| 11 |
Hb_005214_170 |
0.1423830185 |
- |
- |
PREDICTED: uncharacterized protein LOC105636021 [Jatropha curcas] |
| 12 |
Hb_086122_010 |
0.1432891069 |
- |
- |
PREDICTED: immunoglobulin-like domain-containing receptor 1 [Jatropha curcas] |
| 13 |
Hb_005656_170 |
0.1439207171 |
- |
- |
PREDICTED: uncharacterized protein LOC105637447 isoform X1 [Jatropha curcas] |
| 14 |
Hb_017193_010 |
0.1446673584 |
- |
- |
PREDICTED: lactation elevated protein 1 isoform X1 [Jatropha curcas] |
| 15 |
Hb_000212_250 |
0.144786986 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_000331_570 |
0.1464132863 |
- |
- |
PREDICTED: DNA-(apurinic or apyrimidinic site) lyase [Jatropha curcas] |
| 17 |
Hb_006588_190 |
0.1466982881 |
- |
- |
PREDICTED: ataxin-3 homolog [Jatropha curcas] |
| 18 |
Hb_006445_020 |
0.1470247074 |
- |
- |
hypothetical protein L484_026741 [Morus notabilis] |
| 19 |
Hb_000152_180 |
0.1483921431 |
- |
- |
PREDICTED: CDGSH iron-sulfur domain-containing protein NEET [Jatropha curcas] |
| 20 |
Hb_011476_030 |
0.1489351296 |
- |
- |
PREDICTED: uncharacterized protein LOC105632352 [Jatropha curcas] |