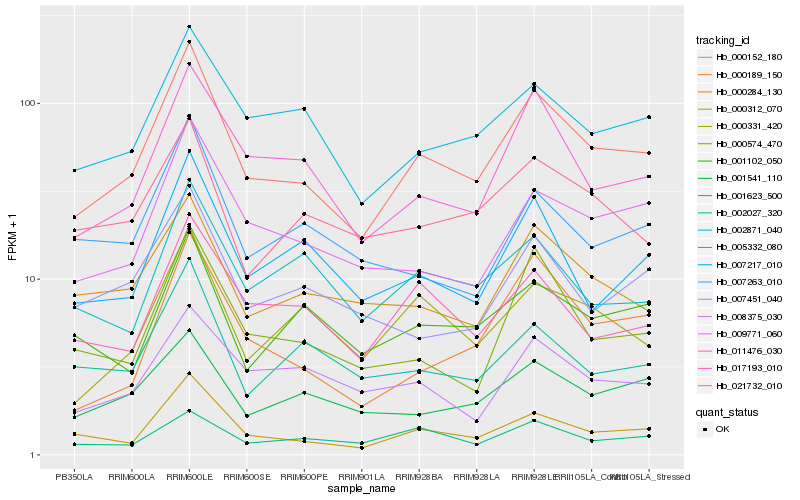
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000152_180 |
0.0 |
- |
- |
PREDICTED: CDGSH iron-sulfur domain-containing protein NEET [Jatropha curcas] |
| 2 |
Hb_000312_070 |
0.0965178384 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_011476_030 |
0.1125584573 |
- |
- |
PREDICTED: uncharacterized protein LOC105632352 [Jatropha curcas] |
| 4 |
Hb_005332_080 |
0.1184683612 |
- |
- |
PREDICTED: succinate dehydrogenase [ubiquinone] iron-sulfur subunit 2, mitochondrial [Jatropha curcas] |
| 5 |
Hb_009771_060 |
0.1206534273 |
- |
- |
PREDICTED: 30S ribosomal protein S31, chloroplastic [Populus euphratica] |
| 6 |
Hb_001541_110 |
0.1275290414 |
- |
- |
PREDICTED: sufE-like protein 2, chloroplastic isoform X2 [Jatropha curcas] |
| 7 |
Hb_007217_010 |
0.1278122975 |
- |
- |
PREDICTED: glycerol-3-phosphate acyltransferase, chloroplastic [Jatropha curcas] |
| 8 |
Hb_000284_130 |
0.1295443735 |
- |
- |
sodium-bile acid cotransporter, putative [Ricinus communis] |
| 9 |
Hb_017193_010 |
0.1296548481 |
- |
- |
PREDICTED: lactation elevated protein 1 isoform X1 [Jatropha curcas] |
| 10 |
Hb_000574_470 |
0.1315995374 |
- |
- |
PREDICTED: chaperone protein dnaJ 1, mitochondrial isoform X1 [Jatropha curcas] |
| 11 |
Hb_000189_150 |
0.1321275666 |
transcription factor |
TF Family: bZIP |
PREDICTED: light-inducible protein CPRF2 [Jatropha curcas] |
| 12 |
Hb_000331_420 |
0.1323962361 |
- |
- |
PREDICTED: nucleoside diphosphate kinase 2, chloroplastic [Jatropha curcas] |
| 13 |
Hb_021732_010 |
0.1371942037 |
- |
- |
biotin carboxylase [Vernicia fordii] |
| 14 |
Hb_001623_500 |
0.1380688115 |
- |
- |
PREDICTED: uncharacterized protein LOC105638487 [Jatropha curcas] |
| 15 |
Hb_007451_040 |
0.1386291665 |
- |
- |
PREDICTED: uncharacterized protein LOC105643411 [Jatropha curcas] |
| 16 |
Hb_008375_030 |
0.1393717367 |
- |
- |
PREDICTED: carbon catabolite repressor protein 4 homolog 4 isoform X1 [Populus euphratica] |
| 17 |
Hb_002027_320 |
0.1406443756 |
- |
- |
PREDICTED: RNA-binding protein BRN1 [Jatropha curcas] |
| 18 |
Hb_001102_050 |
0.1407277098 |
- |
- |
PREDICTED: fatty-acid-binding protein 3 [Jatropha curcas] |
| 19 |
Hb_002871_040 |
0.1415529568 |
- |
- |
PREDICTED: enoyl-[acyl-carrier-protein] reductase [NADH], chloroplastic-like [Populus euphratica] |
| 20 |
Hb_007263_010 |
0.1440106972 |
- |
- |
PREDICTED: clustered mitochondria protein homolog [Jatropha curcas] |