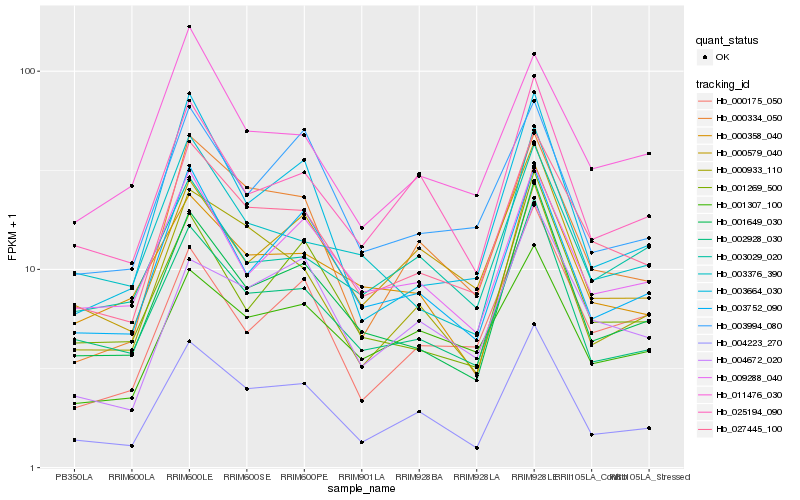
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_027445_100 |
0.0 |
- |
- |
PREDICTED: probable monodehydroascorbate reductase, cytoplasmic isoform 2 [Jatropha curcas] |
| 2 |
Hb_001649_030 |
0.0962488734 |
- |
- |
PREDICTED: peptide deformylase 1B, chloroplastic [Jatropha curcas] |
| 3 |
Hb_000334_050 |
0.098015276 |
- |
- |
PREDICTED: vesicle-associated protein 4-1-like [Jatropha curcas] |
| 4 |
Hb_002928_030 |
0.0989578329 |
- |
- |
hypothetical protein CICLE_v10001788mg [Citrus clementina] |
| 5 |
Hb_011476_030 |
0.1012601051 |
- |
- |
PREDICTED: uncharacterized protein LOC105632352 [Jatropha curcas] |
| 6 |
Hb_004223_270 |
0.1016164691 |
- |
- |
PREDICTED: uncharacterized protein LOC105635371 isoform X1 [Jatropha curcas] |
| 7 |
Hb_009288_040 |
0.1067422851 |
- |
- |
PREDICTED: uncharacterized protein LOC105648677 [Jatropha curcas] |
| 8 |
Hb_001307_100 |
0.1068121872 |
- |
- |
calmodulin binding protein, putative [Ricinus communis] |
| 9 |
Hb_000933_110 |
0.1095341364 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 3 [Jatropha curcas] |
| 10 |
Hb_003752_090 |
0.1099528767 |
- |
- |
chitinase, putative [Ricinus communis] |
| 11 |
Hb_025194_090 |
0.1120924241 |
- |
- |
coproporphyrinogen III oxidase, putative [Ricinus communis] |
| 12 |
Hb_000358_040 |
0.1167186193 |
- |
- |
PREDICTED: probable plastid-lipid-associated protein 4, chloroplastic [Jatropha curcas] |
| 13 |
Hb_003994_080 |
0.1167457196 |
- |
- |
PREDICTED: uncharacterized protein LOC105634384 [Jatropha curcas] |
| 14 |
Hb_000579_040 |
0.1172215991 |
- |
- |
PREDICTED: putative GTP-binding protein 6 [Jatropha curcas] |
| 15 |
Hb_003376_390 |
0.1183969278 |
- |
- |
PREDICTED: 29 kDa ribonucleoprotein A, chloroplastic [Jatropha curcas] |
| 16 |
Hb_004672_020 |
0.1198773153 |
- |
- |
PREDICTED: ATP-dependent zinc metalloprotease FtsH [Jatropha curcas] |
| 17 |
Hb_003029_020 |
0.1207013512 |
- |
- |
ATP-dependent Clp protease proteolytic subunit, putative [Ricinus communis] |
| 18 |
Hb_003664_030 |
0.1223429955 |
- |
- |
PREDICTED: mitochondrial carnitine/acylcarnitine carrier-like protein [Jatropha curcas] |
| 19 |
Hb_000175_050 |
0.1233365918 |
- |
- |
- |
| 20 |
Hb_001269_500 |
0.1265407927 |
- |
- |
PREDICTED: protein LOW PSII ACCUMULATION 3, chloroplastic-like isoform X2 [Populus euphratica] |