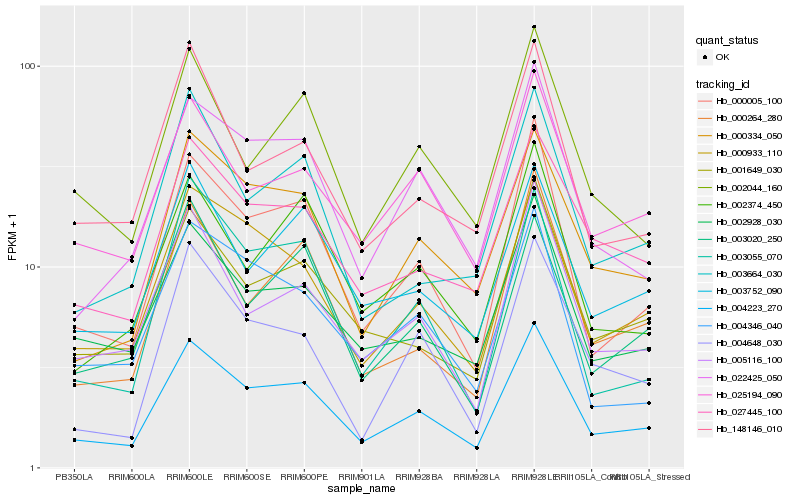
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004223_270 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105635371 isoform X1 [Jatropha curcas] |
| 2 |
Hb_000005_100 |
0.0641205043 |
- |
- |
PREDICTED: pheophorbide a oxygenase, chloroplastic [Jatropha curcas] |
| 3 |
Hb_000933_110 |
0.0830194516 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 3 [Jatropha curcas] |
| 4 |
Hb_000334_050 |
0.0871898019 |
- |
- |
PREDICTED: vesicle-associated protein 4-1-like [Jatropha curcas] |
| 5 |
Hb_022425_050 |
0.0878306162 |
- |
- |
PREDICTED: probable lactoylglutathione lyase, chloroplast isoform X1 [Jatropha curcas] |
| 6 |
Hb_001649_030 |
0.0945009279 |
- |
- |
PREDICTED: peptide deformylase 1B, chloroplastic [Jatropha curcas] |
| 7 |
Hb_005116_100 |
0.0951989839 |
- |
- |
PREDICTED: uncharacterized protein LOC105649999 [Jatropha curcas] |
| 8 |
Hb_002928_030 |
0.0964431764 |
- |
- |
hypothetical protein CICLE_v10001788mg [Citrus clementina] |
| 9 |
Hb_025194_090 |
0.0980583111 |
- |
- |
coproporphyrinogen III oxidase, putative [Ricinus communis] |
| 10 |
Hb_027445_100 |
0.1016164691 |
- |
- |
PREDICTED: probable monodehydroascorbate reductase, cytoplasmic isoform 2 [Jatropha curcas] |
| 11 |
Hb_000264_280 |
0.105050175 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RMA1H1-like [Jatropha curcas] |
| 12 |
Hb_003752_090 |
0.105775433 |
- |
- |
chitinase, putative [Ricinus communis] |
| 13 |
Hb_002044_160 |
0.1059956481 |
- |
- |
PREDICTED: glyoxylate/succinic semialdehyde reductase 1 [Jatropha curcas] |
| 14 |
Hb_002374_450 |
0.1072096563 |
- |
- |
PREDICTED: chlorophyllide a oxygenase, chloroplastic [Jatropha curcas] |
| 15 |
Hb_148146_010 |
0.1095443336 |
- |
- |
coproporphyrinogen III oxidase [Ziziphus jujuba] |
| 16 |
Hb_003664_030 |
0.1104847103 |
- |
- |
PREDICTED: mitochondrial carnitine/acylcarnitine carrier-like protein [Jatropha curcas] |
| 17 |
Hb_003055_070 |
0.1113899035 |
- |
- |
PREDICTED: katanin p80 WD40 repeat-containing subunit B1 homolog isoform X1 [Jatropha curcas] |
| 18 |
Hb_004346_040 |
0.1117694263 |
- |
- |
hypothetical protein POPTR_0016s06630g [Populus trichocarpa] |
| 19 |
Hb_003020_250 |
0.1122769877 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase PP1 isozyme 2 [Jatropha curcas] |
| 20 |
Hb_004648_030 |
0.1150387374 |
- |
- |
PREDICTED: probable dimethyladenosine transferase [Jatropha curcas] |