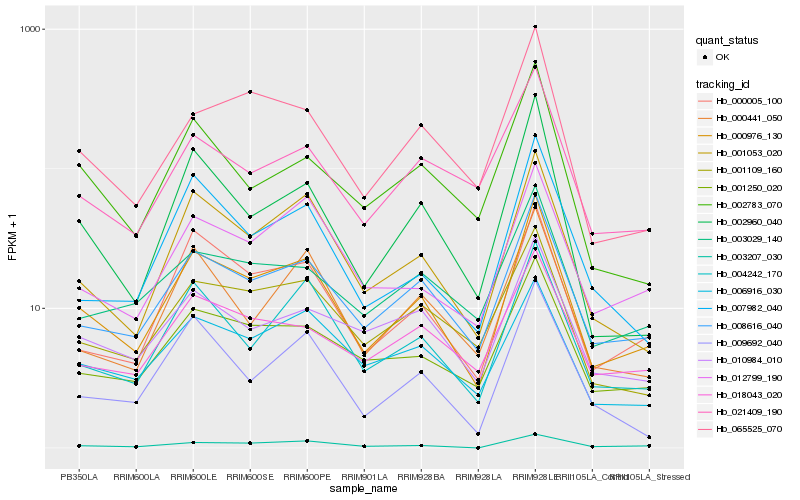
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000976_130 |
0.0 |
- |
- |
PREDICTED: probable BOI-related E3 ubiquitin-protein ligase 2 [Jatropha curcas] |
| 2 |
Hb_008616_040 |
0.0791421211 |
- |
- |
PREDICTED: RNA-binding protein CP33, chloroplastic [Jatropha curcas] |
| 3 |
Hb_001053_020 |
0.0963687204 |
- |
- |
carotenoid cleavage dioxygenase 1 [Manihot esculenta] |
| 4 |
Hb_000005_100 |
0.1065730827 |
- |
- |
PREDICTED: pheophorbide a oxygenase, chloroplastic [Jatropha curcas] |
| 5 |
Hb_001250_020 |
0.1080208719 |
- |
- |
PREDICTED: CRS2-associated factor 2, chloroplastic [Jatropha curcas] |
| 6 |
Hb_004242_170 |
0.1116723376 |
- |
- |
calcium/calmodulin-regulated receptor-like kinase [Manihot esculenta] |
| 7 |
Hb_012799_190 |
0.1163894023 |
- |
- |
PREDICTED: uncharacterized protein LOC105648490 [Jatropha curcas] |
| 8 |
Hb_003207_030 |
0.117138114 |
- |
- |
hypothetical protein VITISV_027754 [Vitis vinifera] |
| 9 |
Hb_001109_160 |
0.11761897 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 10 |
Hb_002960_040 |
0.1202800979 |
- |
- |
PREDICTED: endoplasmin homolog [Jatropha curcas] |
| 11 |
Hb_065525_070 |
0.1208621601 |
- |
- |
unnamed protein product [Homo sapiens] |
| 12 |
Hb_006916_030 |
0.1255392062 |
- |
- |
PREDICTED: uncharacterized protein LOC105646760 [Jatropha curcas] |
| 13 |
Hb_007982_040 |
0.1270862786 |
- |
- |
PREDICTED: uncharacterized aarF domain-containing protein kinase At1g79600, chloroplastic [Jatropha curcas] |
| 14 |
Hb_018043_020 |
0.1277380817 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein MRL1, chloroplastic [Jatropha curcas] |
| 15 |
Hb_021409_190 |
0.1285345053 |
- |
- |
PREDICTED: luminal-binding protein 5-like [Jatropha curcas] |
| 16 |
Hb_010984_010 |
0.1289039004 |
- |
- |
PREDICTED: translation factor GUF1 homolog, chloroplastic isoform X1 [Jatropha curcas] |
| 17 |
Hb_003029_140 |
0.1294235329 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g31400, chloroplastic [Jatropha curcas] |
| 18 |
Hb_009692_040 |
0.1313347405 |
- |
- |
PREDICTED: peptide chain release factor PrfB3, chloroplastic isoform X1 [Jatropha curcas] |
| 19 |
Hb_000441_050 |
0.1326942313 |
- |
- |
PREDICTED: 187-kDa microtubule-associated protein AIR9 [Jatropha curcas] |
| 20 |
Hb_002783_070 |
0.1351906608 |
- |
- |
PREDICTED: endoplasmin homolog [Jatropha curcas] |