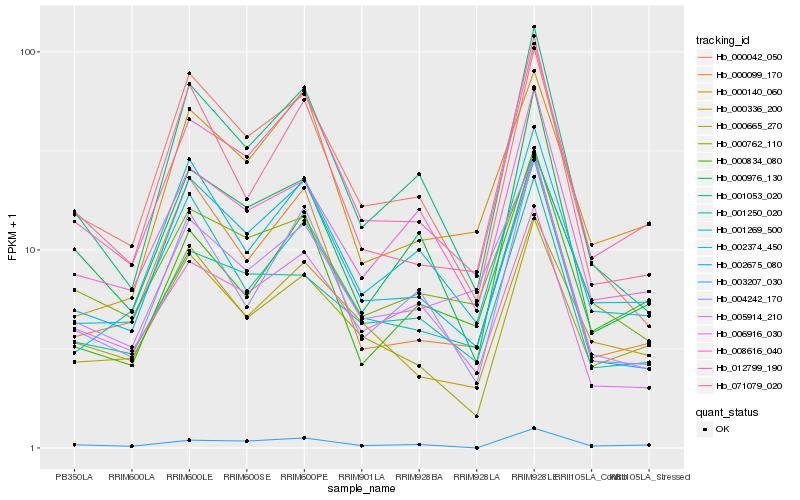
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_012799_190 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105648490 [Jatropha curcas] |
| 2 |
Hb_004242_170 |
0.1062473701 |
- |
- |
calcium/calmodulin-regulated receptor-like kinase [Manihot esculenta] |
| 3 |
Hb_000834_080 |
0.1072589311 |
- |
- |
PREDICTED: uncharacterized protein LOC105637718 [Jatropha curcas] |
| 4 |
Hb_001053_020 |
0.1126265698 |
- |
- |
carotenoid cleavage dioxygenase 1 [Manihot esculenta] |
| 5 |
Hb_000336_200 |
0.1140128183 |
- |
- |
PREDICTED: UPF0481 protein At3g47200-like [Jatropha curcas] |
| 6 |
Hb_000976_130 |
0.1163894023 |
- |
- |
PREDICTED: probable BOI-related E3 ubiquitin-protein ligase 2 [Jatropha curcas] |
| 7 |
Hb_000762_110 |
0.1167644693 |
- |
- |
PREDICTED: protease Do-like 1, chloroplastic [Jatropha curcas] |
| 8 |
Hb_008616_040 |
0.1176811102 |
- |
- |
PREDICTED: RNA-binding protein CP33, chloroplastic [Jatropha curcas] |
| 9 |
Hb_003207_030 |
0.1178465943 |
- |
- |
hypothetical protein VITISV_027754 [Vitis vinifera] |
| 10 |
Hb_001269_500 |
0.1183752695 |
- |
- |
PREDICTED: protein LOW PSII ACCUMULATION 3, chloroplastic-like isoform X2 [Populus euphratica] |
| 11 |
Hb_006916_030 |
0.1186079587 |
- |
- |
PREDICTED: uncharacterized protein LOC105646760 [Jatropha curcas] |
| 12 |
Hb_001250_020 |
0.1196741435 |
- |
- |
PREDICTED: CRS2-associated factor 2, chloroplastic [Jatropha curcas] |
| 13 |
Hb_005914_210 |
0.1214442064 |
- |
- |
PREDICTED: uncharacterized protein LOC105638411 [Jatropha curcas] |
| 14 |
Hb_071079_020 |
0.1219556298 |
- |
- |
RNA polymerase sigma factor rpoD, putative [Ricinus communis] |
| 15 |
Hb_000140_060 |
0.1220406056 |
- |
- |
50S ribosomal protein L5, putative [Ricinus communis] |
| 16 |
Hb_000665_270 |
0.1221429671 |
- |
- |
PREDICTED: adenine phosphoribosyltransferase 5 [Jatropha curcas] |
| 17 |
Hb_002675_080 |
0.1224783006 |
- |
- |
PREDICTED: uncharacterized protein LOC105634950 [Jatropha curcas] |
| 18 |
Hb_000099_170 |
0.1237692487 |
- |
- |
PREDICTED: uncharacterized protein LOC105645733 [Jatropha curcas] |
| 19 |
Hb_002374_450 |
0.1272062523 |
- |
- |
PREDICTED: chlorophyllide a oxygenase, chloroplastic [Jatropha curcas] |
| 20 |
Hb_000042_050 |
0.1318044694 |
- |
- |
PREDICTED: uncharacterized protein LOC105632791 isoform X1 [Jatropha curcas] |