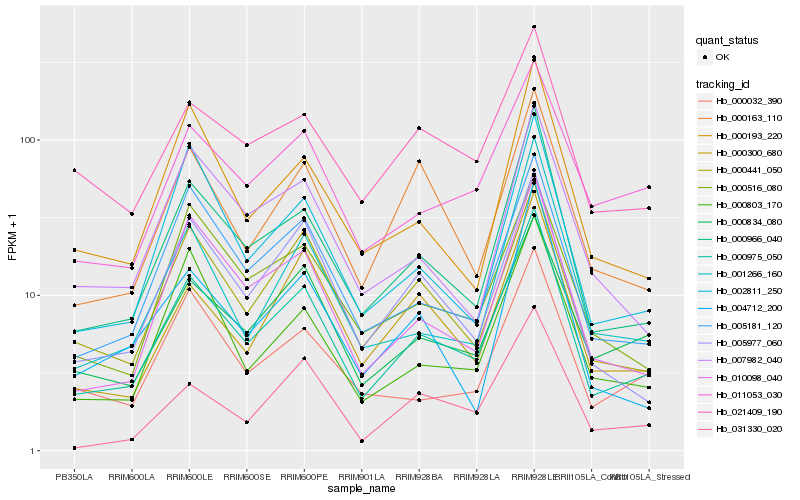
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000975_050 |
0.0 |
- |
- |
PREDICTED: inner membrane protein PPF-1, chloroplastic [Jatropha curcas] |
| 2 |
Hb_000966_040 |
0.0688616625 |
- |
- |
hypothetical protein JCGZ_01642 [Jatropha curcas] |
| 3 |
Hb_010098_040 |
0.1061787459 |
- |
- |
PREDICTED: DNA-binding protein SMUBP-2 [Jatropha curcas] |
| 4 |
Hb_000834_080 |
0.1111375687 |
- |
- |
PREDICTED: uncharacterized protein LOC105637718 [Jatropha curcas] |
| 5 |
Hb_000441_050 |
0.1120193079 |
- |
- |
PREDICTED: 187-kDa microtubule-associated protein AIR9 [Jatropha curcas] |
| 6 |
Hb_005181_120 |
0.1128157939 |
- |
- |
PREDICTED: psbB mRNA maturation factor Mbb1, chloroplastic isoform X1 [Jatropha curcas] |
| 7 |
Hb_005977_060 |
0.1137465598 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_000300_680 |
0.1178323684 |
- |
- |
PREDICTED: uncharacterized protein LOC105632625 isoform X1 [Jatropha curcas] |
| 9 |
Hb_000193_220 |
0.122718152 |
- |
- |
PREDICTED: ribulose-phosphate 3-epimerase, chloroplastic [Jatropha curcas] |
| 10 |
Hb_011053_030 |
0.1229182625 |
- |
- |
- |
| 11 |
Hb_021409_190 |
0.1246184555 |
- |
- |
PREDICTED: luminal-binding protein 5-like [Jatropha curcas] |
| 12 |
Hb_001266_160 |
0.1249888244 |
- |
- |
PREDICTED: pheophytinase, chloroplastic [Jatropha curcas] |
| 13 |
Hb_000032_390 |
0.1266732266 |
- |
- |
PREDICTED: uncharacterized protein LOC105645663 [Jatropha curcas] |
| 14 |
Hb_031330_020 |
0.1268144845 |
- |
- |
PREDICTED: protein IQ-DOMAIN 1-like [Jatropha curcas] |
| 15 |
Hb_004712_200 |
0.1275015077 |
- |
- |
hypothetical protein CICLE_v10010292mg [Citrus clementina] |
| 16 |
Hb_000163_110 |
0.1288004491 |
- |
- |
PREDICTED: blue-light photoreceptor PHR2 [Jatropha curcas] |
| 17 |
Hb_000803_170 |
0.1289929422 |
- |
- |
PREDICTED: uncharacterized protein LOC105648312 [Jatropha curcas] |
| 18 |
Hb_007982_040 |
0.1293724769 |
- |
- |
PREDICTED: uncharacterized aarF domain-containing protein kinase At1g79600, chloroplastic [Jatropha curcas] |
| 19 |
Hb_000516_080 |
0.1293815841 |
- |
- |
hypothetical protein POPTR_0010s24810g [Populus trichocarpa] |
| 20 |
Hb_002811_250 |
0.129762973 |
- |
- |
PREDICTED: probable ribose-5-phosphate isomerase 3, chloroplastic [Jatropha curcas] |