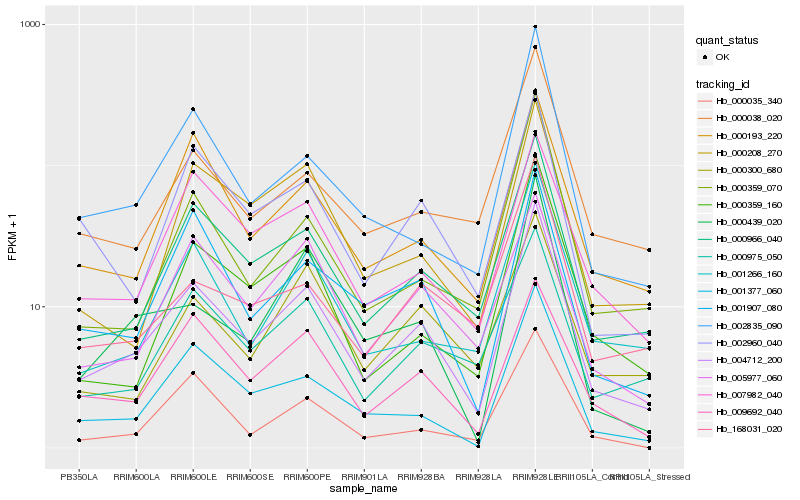
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004712_200 |
0.0 |
- |
- |
hypothetical protein CICLE_v10010292mg [Citrus clementina] |
| 2 |
Hb_000966_040 |
0.0976909081 |
- |
- |
hypothetical protein JCGZ_01642 [Jatropha curcas] |
| 3 |
Hb_001377_060 |
0.1230879219 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g01030, mitochondrial [Jatropha curcas] |
| 4 |
Hb_000975_050 |
0.1275015077 |
- |
- |
PREDICTED: inner membrane protein PPF-1, chloroplastic [Jatropha curcas] |
| 5 |
Hb_001266_160 |
0.1315211761 |
- |
- |
PREDICTED: pheophytinase, chloroplastic [Jatropha curcas] |
| 6 |
Hb_009692_040 |
0.1347814091 |
- |
- |
PREDICTED: peptide chain release factor PrfB3, chloroplastic isoform X1 [Jatropha curcas] |
| 7 |
Hb_000193_220 |
0.1354142891 |
- |
- |
PREDICTED: ribulose-phosphate 3-epimerase, chloroplastic [Jatropha curcas] |
| 8 |
Hb_000359_160 |
0.1374484436 |
- |
- |
PREDICTED: protein CAJ1 [Jatropha curcas] |
| 9 |
Hb_002960_040 |
0.1383909995 |
- |
- |
PREDICTED: endoplasmin homolog [Jatropha curcas] |
| 10 |
Hb_001907_080 |
0.1389092232 |
- |
- |
PREDICTED: uncharacterized protein LOC105628749 [Jatropha curcas] |
| 11 |
Hb_002835_090 |
0.1430799698 |
- |
- |
RecName: Full=Elongation factor Tu, chloroplastic; Short=EF-Tu; Flags: Precursor [Glycine max] |
| 12 |
Hb_000038_020 |
0.1454714938 |
- |
- |
PREDICTED: ATP-dependent zinc metalloprotease FTSH 2, chloroplastic [Jatropha curcas] |
| 13 |
Hb_005977_060 |
0.1455509348 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_007982_040 |
0.1460336382 |
- |
- |
PREDICTED: uncharacterized aarF domain-containing protein kinase At1g79600, chloroplastic [Jatropha curcas] |
| 15 |
Hb_000035_340 |
0.1460715962 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_000208_270 |
0.1461372955 |
- |
- |
PREDICTED: malate dehydrogenase [NADP], chloroplastic [Jatropha curcas] |
| 17 |
Hb_000300_680 |
0.1489514977 |
- |
- |
PREDICTED: uncharacterized protein LOC105632625 isoform X1 [Jatropha curcas] |
| 18 |
Hb_000359_070 |
0.1489664992 |
- |
- |
PREDICTED: ATP-dependent zinc metalloprotease FTSH 2, chloroplastic [Jatropha curcas] |
| 19 |
Hb_000439_020 |
0.149431784 |
transcription factor |
TF Family: ERF |
PREDICTED: dehydration-responsive element-binding protein 3-like [Jatropha curcas] |
| 20 |
Hb_168031_020 |
0.1504576941 |
- |
- |
PREDICTED: uncharacterized protein LOC105646684 isoform X2 [Jatropha curcas] |