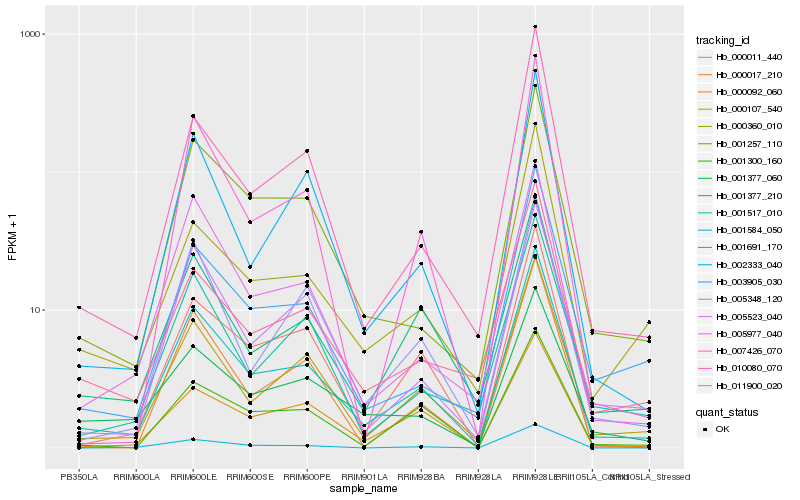
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001584_050 |
0.0 |
- |
- |
PREDICTED: NHL repeat-containing protein 2 isoform X1 [Jatropha curcas] |
| 2 |
Hb_000107_540 |
0.0872622132 |
- |
- |
PREDICTED: uncharacterized protein LOC105635335 [Jatropha curcas] |
| 3 |
Hb_010080_070 |
0.1061432182 |
- |
- |
PREDICTED: ATP-dependent zinc metalloprotease FTSH, chloroplastic [Jatropha curcas] |
| 4 |
Hb_001377_210 |
0.1102975403 |
- |
- |
PREDICTED: clustered mitochondria protein homolog [Jatropha curcas] |
| 5 |
Hb_002333_040 |
0.1125375065 |
- |
- |
hypothetical protein CISIN_1g015895mg [Citrus sinensis] |
| 6 |
Hb_001257_110 |
0.1131554659 |
- |
- |
PREDICTED: uncharacterized protein LOC105636396 [Jatropha curcas] |
| 7 |
Hb_000011_440 |
0.1142102599 |
- |
- |
PREDICTED: GDSL esterase/lipase 1 [Populus euphratica] |
| 8 |
Hb_005977_040 |
0.1183063545 |
- |
- |
Photosystem I reaction center subunit IV A [Medicago truncatula] |
| 9 |
Hb_011900_020 |
0.120172522 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g16390, chloroplastic [Jatropha curcas] |
| 10 |
Hb_007426_070 |
0.1203020817 |
- |
- |
PREDICTED: peroxisomal (S)-2-hydroxy-acid oxidase [Jatropha curcas] |
| 11 |
Hb_005523_040 |
0.1207426157 |
- |
- |
PREDICTED: uncharacterized protein LOC100253680 [Vitis vinifera] |
| 12 |
Hb_003905_030 |
0.1211360353 |
- |
- |
PREDICTED: clustered mitochondria protein homolog [Jatropha curcas] |
| 13 |
Hb_000092_060 |
0.1236224351 |
- |
- |
PREDICTED: uncharacterized protein LOC105646505 [Jatropha curcas] |
| 14 |
Hb_001517_010 |
0.1249830021 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_005348_120 |
0.1269006283 |
- |
- |
PREDICTED: chlorophyll a-b binding protein, chloroplastic [Jatropha curcas] |
| 16 |
Hb_000360_010 |
0.1280153714 |
- |
- |
PREDICTED: ferredoxin-dependent glutamate synthase, chloroplastic isoform X1 [Jatropha curcas] |
| 17 |
Hb_001377_060 |
0.1283175433 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g01030, mitochondrial [Jatropha curcas] |
| 18 |
Hb_001691_170 |
0.1292437233 |
- |
- |
PREDICTED: callose synthase 12-like [Glycine max] |
| 19 |
Hb_001300_160 |
0.129401515 |
- |
- |
PREDICTED: sodium/hydrogen exchanger 2-like [Jatropha curcas] |
| 20 |
Hb_000017_210 |
0.1294077635 |
- |
- |
PREDICTED: homeobox protein BEL1 homolog [Populus euphratica] |