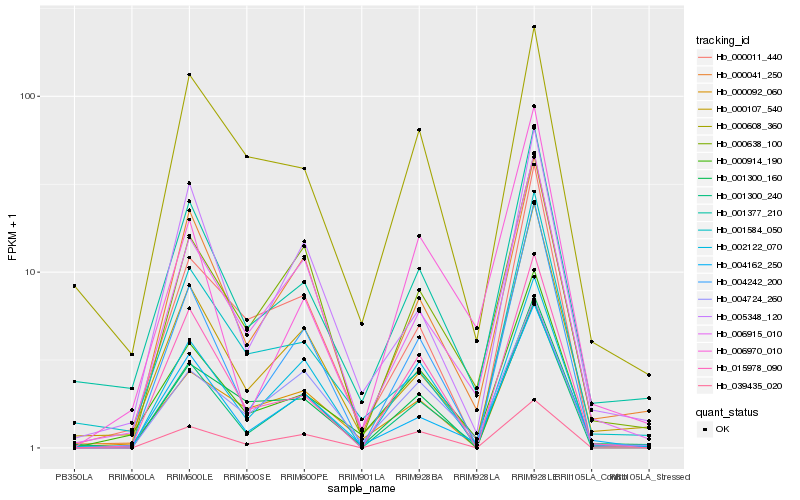
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000914_190 |
0.0 |
- |
- |
xanthine dehydrogenase, putative [Ricinus communis] |
| 2 |
Hb_001377_210 |
0.0980950552 |
- |
- |
PREDICTED: clustered mitochondria protein homolog [Jatropha curcas] |
| 3 |
Hb_000092_060 |
0.1219009848 |
- |
- |
PREDICTED: uncharacterized protein LOC105646505 [Jatropha curcas] |
| 4 |
Hb_004242_200 |
0.1395278646 |
transcription factor |
TF Family: C2H2 |
hypothetical protein POPTR_0001s44130g [Populus trichocarpa] |
| 5 |
Hb_015978_090 |
0.1404090933 |
- |
- |
hypothetical protein POPTR_0014s11700g [Populus trichocarpa] |
| 6 |
Hb_006915_010 |
0.1404834779 |
- |
- |
PREDICTED: clustered mitochondria protein homolog [Jatropha curcas] |
| 7 |
Hb_000011_440 |
0.1421736157 |
- |
- |
PREDICTED: GDSL esterase/lipase 1 [Populus euphratica] |
| 8 |
Hb_000107_540 |
0.1441583704 |
- |
- |
PREDICTED: uncharacterized protein LOC105635335 [Jatropha curcas] |
| 9 |
Hb_001300_160 |
0.1450829013 |
- |
- |
PREDICTED: sodium/hydrogen exchanger 2-like [Jatropha curcas] |
| 10 |
Hb_001300_240 |
0.1472036426 |
- |
- |
shikimate dehydrogenase, putative [Ricinus communis] |
| 11 |
Hb_004724_260 |
0.1499806825 |
- |
- |
PREDICTED: 40S ribosomal protein S14-2-like [Gossypium raimondii] |
| 12 |
Hb_001584_050 |
0.1516219551 |
- |
- |
PREDICTED: NHL repeat-containing protein 2 isoform X1 [Jatropha curcas] |
| 13 |
Hb_000608_360 |
0.152045541 |
- |
- |
calreticulin, putative [Ricinus communis] |
| 14 |
Hb_006970_010 |
0.1551782573 |
- |
- |
lipoxygenase, putative [Ricinus communis] |
| 15 |
Hb_000041_250 |
0.1557248621 |
- |
- |
PREDICTED: uncharacterized protein LOC105641253 [Jatropha curcas] |
| 16 |
Hb_004162_250 |
0.1562509743 |
- |
- |
hypothetical protein POPTR_0004s22660g [Populus trichocarpa] |
| 17 |
Hb_000638_100 |
0.1595175718 |
- |
- |
PREDICTED: histone deacetylase 14 [Jatropha curcas] |
| 18 |
Hb_039435_020 |
0.1631033715 |
- |
- |
Lysosomal Pro-X carboxypeptidase, putative [Ricinus communis] |
| 19 |
Hb_002122_070 |
0.1658011711 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 20 |
Hb_005348_120 |
0.1659093909 |
- |
- |
PREDICTED: chlorophyll a-b binding protein, chloroplastic [Jatropha curcas] |