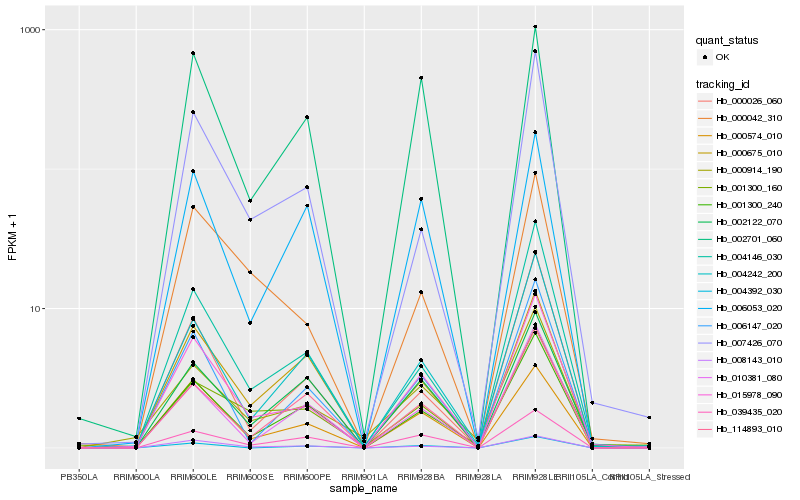
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_015978_090 |
0.0 |
- |
- |
hypothetical protein POPTR_0014s11700g [Populus trichocarpa] |
| 2 |
Hb_008143_010 |
0.1144220817 |
- |
- |
PREDICTED: wall-associated receptor kinase-like 22 [Jatropha curcas] |
| 3 |
Hb_000574_010 |
0.117401583 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein JCGZ_06096 [Jatropha curcas] |
| 4 |
Hb_006147_020 |
0.1215933116 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_004242_200 |
0.1250692363 |
transcription factor |
TF Family: C2H2 |
hypothetical protein POPTR_0001s44130g [Populus trichocarpa] |
| 6 |
Hb_001300_240 |
0.1251695243 |
- |
- |
shikimate dehydrogenase, putative [Ricinus communis] |
| 7 |
Hb_039435_020 |
0.1313972901 |
- |
- |
Lysosomal Pro-X carboxypeptidase, putative [Ricinus communis] |
| 8 |
Hb_004146_030 |
0.1322303862 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |
| 9 |
Hb_000914_190 |
0.1404090933 |
- |
- |
xanthine dehydrogenase, putative [Ricinus communis] |
| 10 |
Hb_000042_310 |
0.142111853 |
- |
- |
Cyclic nucleotide-gated ion channel, putative [Ricinus communis] |
| 11 |
Hb_002122_070 |
0.1439350915 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 12 |
Hb_001300_160 |
0.144056304 |
- |
- |
PREDICTED: sodium/hydrogen exchanger 2-like [Jatropha curcas] |
| 13 |
Hb_002701_060 |
0.1447624486 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_007426_070 |
0.147431643 |
- |
- |
PREDICTED: peroxisomal (S)-2-hydroxy-acid oxidase [Jatropha curcas] |
| 15 |
Hb_004392_030 |
0.1476770754 |
- |
- |
oxidoreductase family protein [Populus trichocarpa] |
| 16 |
Hb_010381_080 |
0.1542509156 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 17 |
Hb_114893_010 |
0.1550761846 |
- |
- |
Flavonol 4'-sulfotransferase, putative [Ricinus communis] |
| 18 |
Hb_000675_010 |
0.1557847262 |
- |
- |
PREDICTED: cytokinin dehydrogenase 7 [Jatropha curcas] |
| 19 |
Hb_006053_020 |
0.156209907 |
- |
- |
CAP [Theobroma cacao] |
| 20 |
Hb_000026_060 |
0.1562249398 |
- |
- |
beta-1,3-glucanase [Manihot esculenta] |