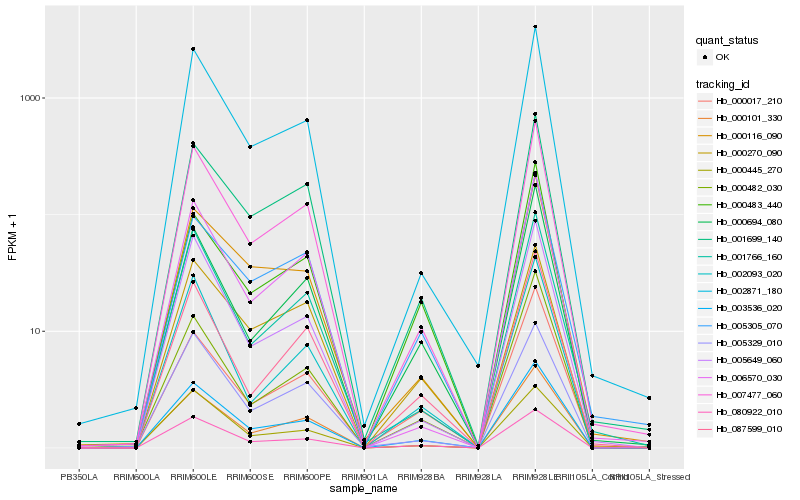
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003536_020 |
0.0 |
- |
- |
PREDICTED: splicing factor 3A subunit 2 [Jatropha curcas] |
| 2 |
Hb_080922_010 |
0.0491461876 |
- |
- |
leucine-rich repeat receptor protein kinase exs precursor, putative [Ricinus communis] |
| 3 |
Hb_007477_060 |
0.0533744719 |
- |
- |
unknown [Medicago truncatula] |
| 4 |
Hb_000101_330 |
0.0639932196 |
- |
- |
- |
| 5 |
Hb_001699_140 |
0.0708691053 |
- |
- |
PREDICTED: ferredoxin--NADP reductase, leaf isozyme, chloroplastic [Jatropha curcas] |
| 6 |
Hb_002871_180 |
0.0725146536 |
- |
- |
photosystem II 10 kDa polypeptide, chloroplastic [Jatropha curcas] |
| 7 |
Hb_000116_090 |
0.0792696716 |
- |
- |
catalase [Hevea brasiliensis] |
| 8 |
Hb_005305_070 |
0.085784497 |
- |
- |
hypothetical protein POPTR_0001s26540g [Populus trichocarpa] |
| 9 |
Hb_087599_010 |
0.0868686777 |
- |
- |
PREDICTED: tropinone reductase homolog [Solanum tuberosum] |
| 10 |
Hb_000694_080 |
0.0876153205 |
- |
- |
phosphoribulose kinase, putative [Ricinus communis] |
| 11 |
Hb_005649_060 |
0.0908397637 |
- |
- |
PREDICTED: uncharacterized protein LOC105644161 [Jatropha curcas] |
| 12 |
Hb_001766_160 |
0.0912001836 |
- |
- |
Serine/threonine-protein kinase SAPK1 [Gossypium arboreum] |
| 13 |
Hb_000017_210 |
0.0913800797 |
- |
- |
PREDICTED: homeobox protein BEL1 homolog [Populus euphratica] |
| 14 |
Hb_002093_020 |
0.0921558272 |
- |
- |
hypothetical protein POPTR_0008s02960g [Populus trichocarpa] |
| 15 |
Hb_000445_270 |
0.0936431989 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 16 |
Hb_006570_030 |
0.0957015022 |
- |
- |
PREDICTED: ferredoxin--NADP reductase, leaf isozyme, chloroplastic [Jatropha curcas] |
| 17 |
Hb_005329_010 |
0.0969627839 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase At1g11410 [Jatropha curcas] |
| 18 |
Hb_000270_090 |
0.0973122094 |
- |
- |
nitrate transporter, putative [Ricinus communis] |
| 19 |
Hb_000482_030 |
0.0981094983 |
transcription factor |
TF Family: C2H2 |
PREDICTED: zinc finger protein NUTCRACKER-like isoform X1 [Populus euphratica] |
| 20 |
Hb_000483_440 |
0.0994659145 |
- |
- |
conserved hypothetical protein [Ricinus communis] |