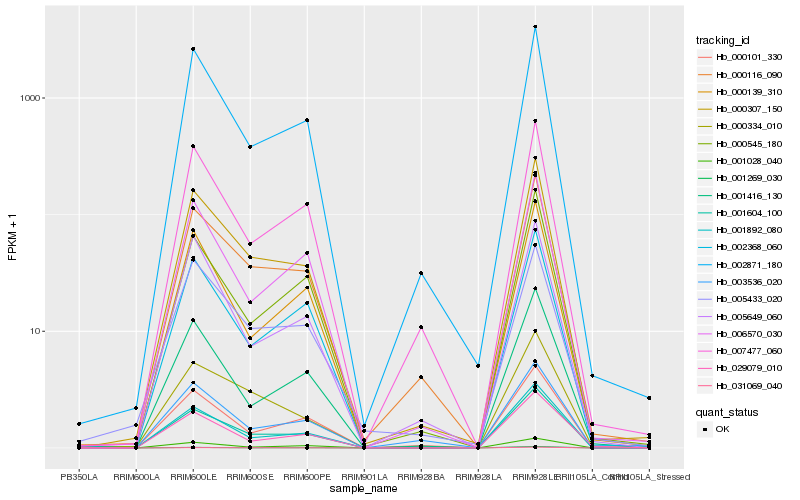
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000307_150 |
0.0 |
- |
- |
PREDICTED: thioredoxin-like protein CDSP32, chloroplastic [Jatropha curcas] |
| 2 |
Hb_000116_090 |
0.0577509628 |
- |
- |
catalase [Hevea brasiliensis] |
| 3 |
Hb_001892_080 |
0.0640186758 |
- |
- |
Clavata3/esr-related 16, putative [Theobroma cacao] |
| 4 |
Hb_002871_180 |
0.0746574642 |
- |
- |
photosystem II 10 kDa polypeptide, chloroplastic [Jatropha curcas] |
| 5 |
Hb_000334_010 |
0.0839521652 |
- |
- |
Cyclic nucleotide-gated ion channel, putative [Ricinus communis] |
| 6 |
Hb_001269_030 |
0.0908954475 |
- |
- |
PREDICTED: pleiotropic drug resistance protein 1-like [Jatropha curcas] |
| 7 |
Hb_007477_060 |
0.0957801082 |
- |
- |
unknown [Medicago truncatula] |
| 8 |
Hb_006570_030 |
0.0962665549 |
- |
- |
PREDICTED: ferredoxin--NADP reductase, leaf isozyme, chloroplastic [Jatropha curcas] |
| 9 |
Hb_001416_130 |
0.096961544 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_001604_100 |
0.0987064433 |
- |
- |
PREDICTED: putative serine/threonine-protein kinase isoform X3 [Jatropha curcas] |
| 11 |
Hb_000139_310 |
0.0997286026 |
- |
- |
PREDICTED: glucose-1-phosphate adenylyltransferase large subunit 1, chloroplastic [Jatropha curcas] |
| 12 |
Hb_000101_330 |
0.0999044461 |
- |
- |
- |
| 13 |
Hb_002368_060 |
0.1003900934 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_005433_020 |
0.1019623472 |
- |
- |
PREDICTED: multicopper oxidase LPR1 [Jatropha curcas] |
| 15 |
Hb_031069_040 |
0.1021356301 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 16 |
Hb_029079_010 |
0.1037596181 |
- |
- |
receptor-kinase, putative [Ricinus communis] |
| 17 |
Hb_003536_020 |
0.1041673456 |
- |
- |
PREDICTED: splicing factor 3A subunit 2 [Jatropha curcas] |
| 18 |
Hb_000545_180 |
0.105474241 |
- |
- |
Photosystem II stability/assembly factor HCF136, chloroplast precursor, putative [Ricinus communis] |
| 19 |
Hb_005649_060 |
0.106571084 |
- |
- |
PREDICTED: uncharacterized protein LOC105644161 [Jatropha curcas] |
| 20 |
Hb_001028_040 |
0.1073595137 |
- |
- |
hypothetical protein POPTR_0018s09860g [Populus trichocarpa] |