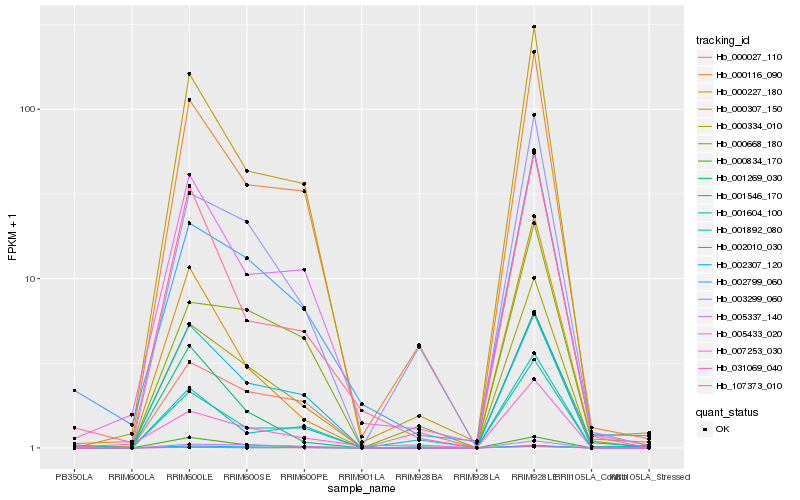
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000334_010 |
0.0 |
- |
- |
Cyclic nucleotide-gated ion channel, putative [Ricinus communis] |
| 2 |
Hb_000307_150 |
0.0839521652 |
- |
- |
PREDICTED: thioredoxin-like protein CDSP32, chloroplastic [Jatropha curcas] |
| 3 |
Hb_000116_090 |
0.0988234911 |
- |
- |
catalase [Hevea brasiliensis] |
| 4 |
Hb_031069_040 |
0.1050921913 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 5 |
Hb_002799_060 |
0.1128775449 |
- |
- |
PREDICTED: zeaxanthin epoxidase, chloroplastic-like [Jatropha curcas] |
| 6 |
Hb_003299_060 |
0.1195884189 |
- |
- |
nitrate transporter, putative [Ricinus communis] |
| 7 |
Hb_001604_100 |
0.1199760273 |
- |
- |
PREDICTED: putative serine/threonine-protein kinase isoform X3 [Jatropha curcas] |
| 8 |
Hb_001892_080 |
0.1339396427 |
- |
- |
Clavata3/esr-related 16, putative [Theobroma cacao] |
| 9 |
Hb_005433_020 |
0.1350636222 |
- |
- |
PREDICTED: multicopper oxidase LPR1 [Jatropha curcas] |
| 10 |
Hb_000834_170 |
0.1354766977 |
- |
- |
polygalacturonase, putative [Ricinus communis] |
| 11 |
Hb_000027_110 |
0.1376059344 |
- |
- |
wall-associated kinase, putative [Ricinus communis] |
| 12 |
Hb_005337_140 |
0.138339345 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: wall-associated receptor kinase 1-like [Jatropha curcas] |
| 13 |
Hb_002010_030 |
0.1399323271 |
- |
- |
Phytosulfokine receptor precursor, putative [Ricinus communis] |
| 14 |
Hb_002307_120 |
0.1405727144 |
- |
- |
scab resistance family protein [Populus trichocarpa] |
| 15 |
Hb_001269_030 |
0.1410309831 |
- |
- |
PREDICTED: pleiotropic drug resistance protein 1-like [Jatropha curcas] |
| 16 |
Hb_000227_180 |
0.1434703543 |
- |
- |
PREDICTED: glucose-1-phosphate adenylyltransferase large subunit 1, chloroplastic [Jatropha curcas] |
| 17 |
Hb_000668_180 |
0.1440075049 |
- |
- |
hypothetical protein POPTR_0005s16490g [Populus trichocarpa] |
| 18 |
Hb_107373_010 |
0.1460142872 |
- |
- |
PREDICTED: zeaxanthin epoxidase, chloroplastic-like [Jatropha curcas] |
| 19 |
Hb_007253_030 |
0.1463476656 |
- |
- |
PREDICTED: 33 kDa ribonucleoprotein, chloroplastic-like [Populus euphratica] |
| 20 |
Hb_001546_170 |
0.1472457606 |
- |
- |
ankyrin repeat-containing protein, putative [Ricinus communis] |