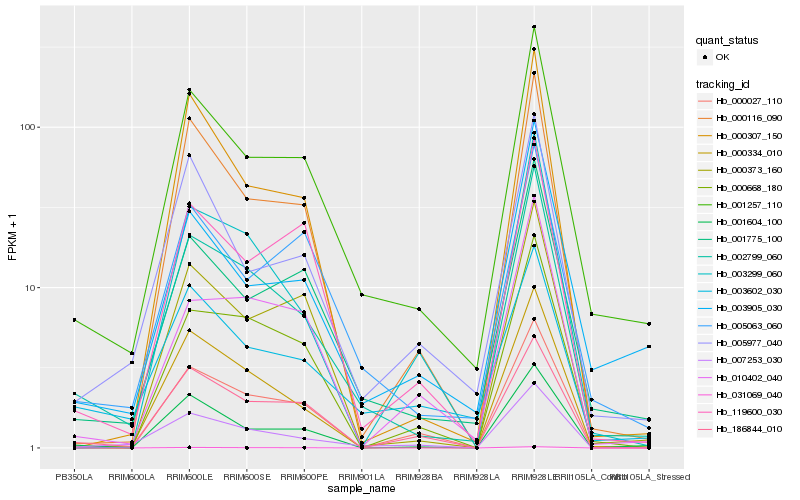
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002799_060 |
0.0 |
- |
- |
PREDICTED: zeaxanthin epoxidase, chloroplastic-like [Jatropha curcas] |
| 2 |
Hb_001257_110 |
0.0988696612 |
- |
- |
PREDICTED: uncharacterized protein LOC105636396 [Jatropha curcas] |
| 3 |
Hb_007253_030 |
0.1125090864 |
- |
- |
PREDICTED: 33 kDa ribonucleoprotein, chloroplastic-like [Populus euphratica] |
| 4 |
Hb_000334_010 |
0.1128775449 |
- |
- |
Cyclic nucleotide-gated ion channel, putative [Ricinus communis] |
| 5 |
Hb_001775_100 |
0.1297175795 |
- |
- |
PREDICTED: probable 2-carboxy-D-arabinitol-1-phosphatase [Jatropha curcas] |
| 6 |
Hb_001604_100 |
0.1367088606 |
- |
- |
PREDICTED: putative serine/threonine-protein kinase isoform X3 [Jatropha curcas] |
| 7 |
Hb_031069_040 |
0.1418899971 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 8 |
Hb_000116_090 |
0.1437562156 |
- |
- |
catalase [Hevea brasiliensis] |
| 9 |
Hb_003299_060 |
0.14391193 |
- |
- |
nitrate transporter, putative [Ricinus communis] |
| 10 |
Hb_000668_180 |
0.1463683149 |
- |
- |
hypothetical protein POPTR_0005s16490g [Populus trichocarpa] |
| 11 |
Hb_000307_150 |
0.1481015361 |
- |
- |
PREDICTED: thioredoxin-like protein CDSP32, chloroplastic [Jatropha curcas] |
| 12 |
Hb_005063_060 |
0.1549308274 |
transcription factor |
TF Family: GNAT |
PREDICTED: uncharacterized protein LOC105647704 [Jatropha curcas] |
| 13 |
Hb_000027_110 |
0.1566982173 |
- |
- |
wall-associated kinase, putative [Ricinus communis] |
| 14 |
Hb_000373_160 |
0.1578309369 |
- |
- |
PREDICTED: fructose-1,6-bisphosphatase, cytosolic [Jatropha curcas] |
| 15 |
Hb_010402_040 |
0.1590280697 |
- |
- |
hypothetical protein JCGZ_21227 [Jatropha curcas] |
| 16 |
Hb_003602_030 |
0.1597117628 |
- |
- |
PREDICTED: putative pentatricopeptide repeat-containing protein At5g52630 [Jatropha curcas] |
| 17 |
Hb_186844_010 |
0.1610506489 |
- |
- |
leucine-rich repeat receptor protein kinase exs precursor, putative [Ricinus communis] |
| 18 |
Hb_119600_030 |
0.1611572876 |
- |
- |
PREDICTED: uncharacterized protein LOC105635187 [Jatropha curcas] |
| 19 |
Hb_003905_030 |
0.1622857596 |
- |
- |
PREDICTED: clustered mitochondria protein homolog [Jatropha curcas] |
| 20 |
Hb_005977_040 |
0.1631834004 |
- |
- |
Photosystem I reaction center subunit IV A [Medicago truncatula] |