| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
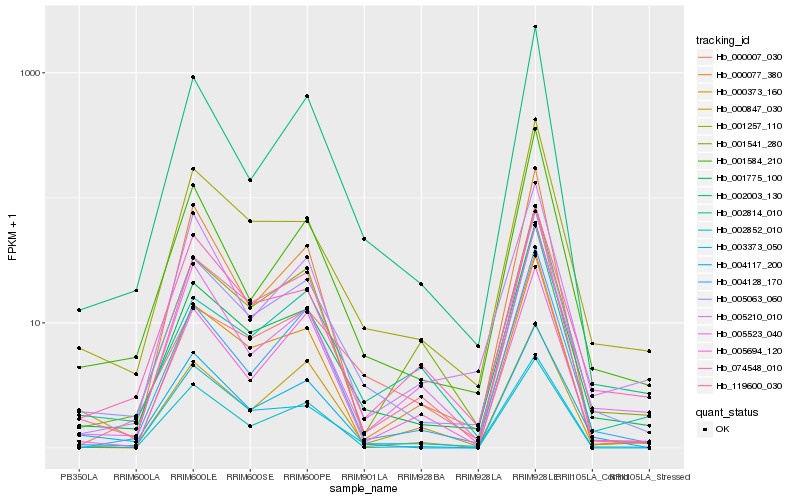
Hb_005063_060 |
0.0 |
transcription factor |
TF Family: GNAT |
PREDICTED: uncharacterized protein LOC105647704 [Jatropha curcas] |
| 2 |
Hb_001775_100 |
0.0667368697 |
- |
- |
PREDICTED: probable 2-carboxy-D-arabinitol-1-phosphatase [Jatropha curcas] |
| 3 |
Hb_001257_110 |
0.0921854153 |
- |
- |
PREDICTED: uncharacterized protein LOC105636396 [Jatropha curcas] |
| 4 |
Hb_002003_130 |
0.0969891961 |
- |
- |
hypothetical protein CISIN_1g027843mg [Citrus sinensis] |
| 5 |
Hb_119600_030 |
0.0999623146 |
- |
- |
PREDICTED: uncharacterized protein LOC105635187 [Jatropha curcas] |
| 6 |
Hb_004117_200 |
0.1095371425 |
- |
- |
PREDICTED: RING-H2 finger protein ATL74 [Jatropha curcas] |
| 7 |
Hb_001584_210 |
0.1124278697 |
- |
- |
PREDICTED: chloroplast stem-loop binding protein of 41 kDa b, chloroplastic [Jatropha curcas] |
| 8 |
Hb_004128_170 |
0.1132901725 |
- |
- |
hypothetical protein POPTR_0016s08350g [Populus trichocarpa] |
| 9 |
Hb_000007_030 |
0.1237086922 |
- |
- |
PREDICTED: chaperonin 60 subunit beta 4, chloroplastic isoform X1 [Jatropha curcas] |
| 10 |
Hb_074548_010 |
0.1247494681 |
- |
- |
PREDICTED: 28 kDa ribonucleoprotein, chloroplastic isoform X1 [Jatropha curcas] |
| 11 |
Hb_005523_040 |
0.1252589578 |
- |
- |
PREDICTED: uncharacterized protein LOC100253680 [Vitis vinifera] |
| 12 |
Hb_005694_120 |
0.1280069701 |
- |
- |
PREDICTED: calcium-transporting ATPase, endoplasmic reticulum-type [Jatropha curcas] |
| 13 |
Hb_000373_160 |
0.1288068863 |
- |
- |
PREDICTED: fructose-1,6-bisphosphatase, cytosolic [Jatropha curcas] |
| 14 |
Hb_002852_010 |
0.1290537331 |
- |
- |
leucine-rich repeat receptor protein kinase exs precursor, putative [Ricinus communis] |
| 15 |
Hb_005210_010 |
0.1302992177 |
- |
- |
PREDICTED: uncharacterized protein LOC105638163 [Jatropha curcas] |
| 16 |
Hb_001541_280 |
0.1303820945 |
- |
- |
phosphate transporter [Manihot esculenta] |
| 17 |
Hb_000077_380 |
0.131721449 |
- |
- |
PREDICTED: protochlorophyllide reductase [Jatropha curcas] |
| 18 |
Hb_002814_010 |
0.1327881723 |
- |
- |
PREDICTED: adenylate kinase 5, chloroplastic [Jatropha curcas] |
| 19 |
Hb_000847_030 |
0.1341714304 |
- |
- |
PREDICTED: asparagine--tRNA ligase, cytoplasmic 2 [Jatropha curcas] |
| 20 |
Hb_003373_050 |
0.1343573708 |
- |
- |
receptor protein kinase zmpk1, putative [Ricinus communis] |